Background
Fish is a major source of food and income globally (Tacon and Metian 2013). A number of countries have been dependent on capture fisheries as the major source of fish for decades. Despite the worldwide increase in fish production, a decline has been observed in several cases (Burkhardt-Holm et al. 2002; Noakes et al. 2000) and this has been majorly attributed to over-fishing and climate-related threats. For Uganda in particular, production dropped from about 0.5 million tons in the year 2007 to only 0.38 million tons in 2010 amidst several efforts by the government to increase production (MAAIF 2012). Indeed, Ogutu-Ohwayo (Ogutu-Ohwayo 1990) reported a decrease in native species of fish in Lakes Victoria and Kyoga, the two major capture fish sources in the country. In the recent past, more emphasis has been put into fish farming worldwide (Tidwell and Allan 2001) and East-Africa in particular (Rutaisire et al. 2009). This has resulted in a significant contribution from aquaculture to the overall fish production (Brander 2007). Oreochromis niloticus (Nile tilapia) and Clarius gariepinus (African catfish) are the two most cultured fish species in Uganda (Akoll and Mwanja 2012). Though still in its infancy, aquaculture is a potential alternative to capture fisheries in Uganda if properly developed. Unfortunately, intensive fish farming is associated with a number of challenges including diseases (Bondad-Reantaso et al. 2005; Walakira et al. 2014), which demotivate farmers due to the increased economic loss.
Bacterial pathogens are a great threat to fish production worldwide due to the high economic importance of diseases they cause (Bondad-Reantaso et al. 2005; Subasinghe et al. 2001). A number of bacterial pathogens have been reported to cause fish diseases worldwide. Bacteria of particular importance in fresh water fish include Streptococcus spp., Aeromonas spp., Flavobacterium spp., Edwardsiella spp., Pseudomonas spp., Vibrio spp., and Mycobacterium spp. (Cipriano 2001; Meyer and Bullock 1973; Abowei and Briyai 2011; Lio-Po and Lim 2014; Mohanty and Sahoo 2007) among others. Infections of the Nile tilapia and African catfish have been reported involving such bacteria in East-Africa especially Kenya (Akoll and Mwanja 2012), with a few reports from Uganda (Walakira et al. 2014).
In Uganda, much of the attention has been given to parasitic diseases (Akoll and Mwanja 2012). A few studies focusing on bacteria were limited to identification of the pathogens at genus level (Walakira et al. 2014). This has left the status of fish bacterial pathogens and the associated diseases in the country improperly understood with very scanty information about their epidemiological parameters. This in addition to the poor disease diagnostics and control measures characteristic of the country’s fisheries sector (Akoll and Mwanja 2012) has led to a likelihood that many fish diseases remain undiagnosed and could continuously affect production.
Knowledge about antibiotic susceptibility of bacteria is vital for the proper management of the diseases they cause. Worldwide, use of antibiotics in aquaculture and the potential transmission of resistant bacteria between terrestrial and aquatic environments have been reported (Cabello 2006). The common mechanism of this transmission is via transfer of R plasmids with fish bacteria acting as intermediate vectors (Sørum 2006). However, several studies have focused on the transmission of antimicrobial resistance (AMR) between humans and terrestrial food animals (Barton 2000; Van den Bogaard and Stobberingh 2000), with less attention to the aquatic ecosystem including fish. This has left inadequate information available about the antimicrobial drug susceptibility of the aquatic ecosystem, a very important aspect in the epidemiology of AMR (Biyela et al. 2004). In Uganda, high levels of antibiotic misuse (Mukonzo et al. 2013); and antibiotic resistance in the human and livestock populations have been reported (Byarugaba 2004; Byarugaba et al. 2011; UNAS 2015). Though the available information suggests minimal use of antibiotics in aquaculture in Uganda, the use of animal wastes to fertilize ponds (Sørum and Sunde 2001) and the close interaction between humans, livestock and the aquatic ecosystem indicates a possible transmission of AMR to the aquatic environment. The widespread occurrence of naturally resistant bacteria in the aquatic environment and soil could also contribute to the passage of antibiotic resistance genes to fish bacteria (Cantas et al. 2013). Besides, high levels of AMR have been reported in bacterial isolates from fish elsewhere (Newaj-Fyzul et al. 2008; Sarter et al. 2007). However, no information is available on AMR for fish bacteria in Uganda amidst glaring reality for the impact of AMR and the increase in intensive aquaculture that could warrant antibiotic use. We, therefore, conducted this study to identify the different bacteria infecting Nile tilapia and African catfish and to determine the antibiotic susceptibility of selected fish bacteria in Uganda.
Methods
This cross section study involved collection of fish samples from five districts of Uganda. Four of the districts including Mukono, Mpigi, Wakiso and Buikwe are from the central region while one (Kole district) is from the northern region (Fig. 1). Bacteria were isolated from the collected fish and identified by morphological and biochemical characteristics. Antibiotic susceptibility of selected bacteria was determined by the Kirby-Bauer disc diffusion method (Hudzicki 2009).
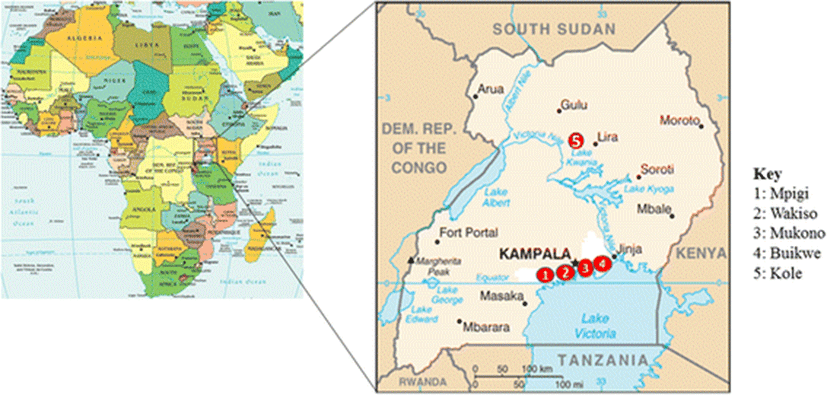
A total of 288 fish with an average of 6 fish per farm were collected from 40 purposively selected fish farms from the following districts; Wakiso (16), Mukono (7), Buikwe (6), Mpigi (6) and Kole (5). The farms included 33 earthen ponds, 5 cages and 2 tanks. It also included 57 fish collected from 8 wild water sites that are majorly landing sites around Lake Victoria (Table 1). The water temperatures ranged between 24.3-28 °C with an average of 25.5 °C for all sites where temperature measurements were taken. The collected fish were immediately transported in cool boxes or buckets containing their source waters, to the Microbiology Laboratory at the College of Veterinary Medicine, Animal Resources and Biosecurity, (CoVAB), Makerere University.
Live fish were humanely killed by a sharp blow to the head. Fish surfaces were disinfected using 70% ethanol. The fish were aseptically dissected to obtain tissue samples of the head-kidney, liver, spleen and the brain. The tissues sampled from individual fish were pooled and homogenized in sterile phosphate buffered saline (PBS) using a mortar and pestle. Bacteriological swabs were taken from any gross lesions observed. The homogenate and swabs were inoculated on Brain Heart Infusion (BHI) agar, Trypticase Soy Agar (TSA) and Blood Agar, in addition to some selective nutrient agar plates including TCBS agar, Shiers agar and Pseudomonas agar, all from Laboratorios CONDA. The plates were incubated between 25 and 30 °C for 24 to 48 h and pure cultures were obtained by sub-culturing.
The isolates were identified following previously described methods (Fish and Service 2010; MacFaddin 2000). In brief, colony morphology including shape, color, pigmentation, haemolytic activity, size, edges and elevation were determined and isolates grouped accordingly. Two (2) to four (4) representative isolates from each group were subjected to Gram staining, motility testing and a battery of biochemical tests including; oxidase, catalase, triple sugar iron (TSI), indole, urea slants, citrate slants, methyl red, oxidative fermentation, nitrate reduction and gelatin liquefaction. The tentatively identified isolates were preserved on agar slants and transported to the Microbiology laboratory of the Faculty of Veterinary Medicine, Norwegian University of Life Sciences (NMBU), Oslo, Norway. Here, further identification of the isolates was achieved using Analytical Profile Index (API) test kits 20E and 20NE (Biomerieux), following the manufacturer’s instruction.
Antibiotic susceptibility testing was carried out by Disc diffusion method according to (Hudzicki 2009). Two genera were tested: genus Aeromonas, due to its high pathogenicity in fish and high prevalence in the study area. It has also been used as an indicator bacterium for antibiotic susceptibility in aquatic environments (Baron et al. 2017; Penders and Stobberingh 2008; Usui et al. 2016); and Genus Plesiomonas due to its high prevalence in the study area, it was considered to represent family Enterobacteriaceae, a group to which many fish bacteria belongs. A total of 155 isolates (Aeromonas spp. =82 and P. shigelloides = 73) were tested against fourteen (14) antibiotics, the majority of which are commonly used not only in human medicine but also the livestock sectors of Uganda. Standard antibiotic disks (Neo-Sensitabs™, Rosco Diagnostica A/S) containing Sulfamethoxazole-trimethoprim combination (25 μg), Penicillin (10 μg), Tetracycline (30 μg), Gentamycin (10 μg), Streptomycin (10 μg), Imipenem (10 μg), Erythromycin (15 μg), Ampicillin (10 μg), Ceftaxime (30 μg), Chloramphenicol (30 μg), Aztreonam (30 μg), Nalidixic acid (30 μg), Enrofloxacin (10 μg), Oxacillin (1 μg) and Ceftriaxone (30 μg), were used on Mueller Hinton agar (Merck KGaA, Germany). In brief, the isolates were plated on Brain Heart Infusion (BHI) agar and incubated for 24 h. Colonies were picked and emulsified in 0.85% Sodium chloride to create a suspension matching 0.5 McFarland standard, an approximate concentration of 1.5 × 108 CFU/ml. Two hundred μl of the suspension was inoculated on the plates, spread using a sterile loop and allowed to dry for 2–5 min. The antibiotic discs were thereafter applied to the plates using a disc dispenser (Neo-Sensitabs™, Rosco Diagnostica A/S) and incubated at 25-30 °C for 18–24 h. The zones of clearance were measured using a plate ruler. The isolates were defined as Resistant (R), Intermediate (I) or Susceptible (S) according to CLSI (CLSI 2006; CLSI 2015). E. coli strain ATCC 25922 and Pseudomonas aeruginosa strain ATCC 27853 were used as controls.
Results
A total of 288 fish samples including Nile tilapia 81.6% (n = 235) and African catfish 18.4% (n = 53) were collected from ponds (68.8%, n = 33), cages (10.4%, n = 5), tanks (4.2%, n = 2) and wild water sites (16.7%, n = 8). Wakiso district had the highest number of the farms sampled (33.3, n = 16) while Kole had the least number (10.4%, n = 5). Details are shown in Table 1.
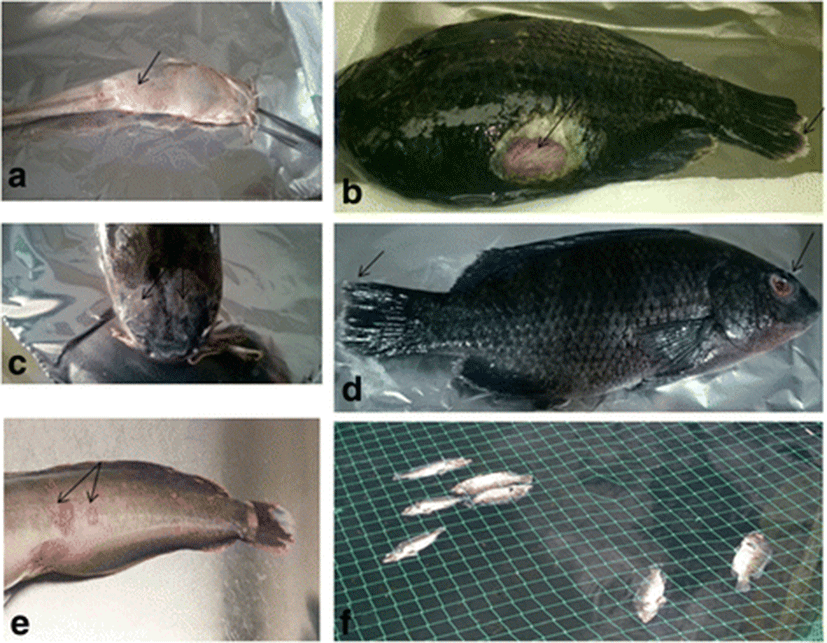
Of the 288 fish collected from all the sites (n = 48), 69 were from diseased farms. This included 46 tilapia from 7 farms and 23 catfish from 4 farms. Common disease signs observed in both tilapia and catfish were skin ulcers, haemorrhagic lesions on the skin, exophthalmos, swollen abdomen and accumulation of serous bloody fluids in the abdomen. Fin rot and stunted growth occurred in tilapia. In addition, mortalities were observed in both tilapia and catfish (Fig. 2). All the fish collected from the wild were apparently healthy.
A total of 15 genera, with at least 18 different species of bacteria were identified. Of particular interest, the following species and their corresponding prevalence at farm level were found: Aeromonas hydrophila (43.8%), Aeromonas sobria (20.8%), Edwardsiella tarda (8.3%), Streptococcus spp. (6.3%) and Flavobacterium spp. (4.2%). Several other bacteria, including those being reported for the first time in fish from Uganda were also isolated. Some of the bacteria isolated from 25% (n = 12) of the sites (farms and wild waters, for both catfish and Nile tilapia) were not properly identified and were presented as ‘unidentified bacteria’. All the 15 genera isolated in this study occurred in Nile tilapia while only 26.7% of them were isolated from African catfish. (Table 2). There was a significant difference (p < 0.05) in the occurrence of the different bacteria between the two types of fish farms.
The table shows the numbers and percentages of Nile tilapia and Catfish farms from where the corresponding bacteria were isolated
All ponds, cages, tanks and wild sites sampled had one or more fish infected by at least one species of bacteria. About 88.9% (n = 16), 61.1% (n = 11), 33% (n = 6) and 11% (n = 2) of all the different genera/ species of bacteria identified (n = 18) occurred in ponds, cages, wild water sites and tanks, respectively, with a significant difference (p < 0.05) in the occurrence of different bacteria across the production systems. Pathogens of great importance in fish including Aeromonas hydrophila, Aeromonas sobria, Edwardsiella tarda, Flavobacterium spp. and Streptococcus spp. were mainly isolated from ponds and cages. Some bacteria such as Ralstonia picketti, Pseudomonas stutzeri and Burkholderia cepacia were very scanty, occurring at only 2.1% of the farms/sites sampled (Table 3).
The table shows the numbers and percentages of the different types of production systems (ponds, cages, wild or tanks) from where the corresponding bacteria were isolated
Aeromonas spp. (n = 82) showed 100% susceptibility to Sulphamethoxazole-trimethoprim, Tetracycline, Gentamycin, Streptomycin, Imipenem, Ceftriaxone, Chloramphenicol, Aztreonam, Nalidixic acid and Enrofloxacin. However, the same isolates were 100% resistant to Penicillin and Ampicillin and about 23.2% were resistant to Ceftaxime. Majority of P. shigelloides were also susceptible to most of the drugs. They however expressed 100% resistance to Penicillin and Oxacillin, with intermediate resistance common to Ampicillin. Both genera expressed varying levels of resistance to Erythromycin (Table 4). There was no significant difference (p < 0.05) in antibiotics susceptibility between the different production systems, and the two species of bacteria tested. Resistance phenotypes were, however, observed more in farmed compared to wild fish.
SXT Sulfamethoxazole-Trimethoprim, PEN Penicillin, TET Tetracycline, GEN Gentamycin, STR Streptomycin, IPM Imipenem, ERY Erythromycin, AMP Ampicillin, CTX Ceftaxime, CTR Ceftriaxone, CLR Chloramphenicol, AZT Aztreonam, NAL Nalidixic acid, ENF Enrofloxacin, OXA Oxacillin, i Intermediate, r Resistant, n number of isolates tested from the different production systems and ‘-‘means no isolate tested
Discussion
Knowledge about prevalent etiological agents and treatment options is necessary for proper management and control of disease in a susceptible population. In this study, we identified the different bacteria infecting O. niloticus and C. gariepinus; and determined the antibiograms of selected fish bacteria in Uganda. A total of 15 different genera of bacteria were isolated. This included bacteria known to cause serious disease in fish and those of varying significance as fish pathogens. Joh and colleagues also reported 15 genera of bacteria in eels from Korea (Joh et al. 2013) with majority of the genera common between the two studies. However, Newaj-Fyzul and others isolated 13 genera from tilapia in Trinidad (Newaj-Fyzul et al. 2008), with only 4 (26.7%) genera common between the two studies.
Among the bacteria identified in the present study are several genera and species being reported for the first time in fish from Uganda. Some of these bacteria are common pathogens known for causing heavy mortalities in fish elsewhere. These include Edwardsiella tarda, the causative agent of edwardsiellosis, an epizootic disease of cultured and wild fish (Mohanty and Sahoo 2007; Wakabayashi 1973), Aeromonas sobria, and Aeromonas hydrophila, which has been isolated from several cases involving highly virulent strains (Hemstreet 2010; Nielsen et al. 2001; Chen and Lu 1991). Aeromonas spp. were the most common bacteria isolated in this study with about 43.8% of the farms infected with A. hydrophila and 20.1% with A. sobria. Indeed, similar findings have been reported from the same study area (Walakira et al. 2014) and elsewhere (Joh et al. 2013). Other researchers have also reported similar findings (Newaj-Fyzul et al. 2008; Ribeiro et al. 2010) and this could be attributed to the ubiquitous nature of Aeromonas spp. in the aquatic environment, similarity of production systems and fish species involved in the different studies.
The prevalence of E. tarda (8.3%) and Flavobacterium spp. (4.2%) were generally low. This shows they were not major contributors to the clinical disease encountered in this study. Similar findings have been reported by other researchers (Newaj-Fyzul et al. 2008; Joh et al. 2013). However, some studies reported the prevalence of Flavobacterium spp. to be as high as 35% (Walakira et al. 2014) and 55.6% (Tamale et al. 2011) respectively, moreover from the same study area as the current study. These differences could be due to variation in seasons and sensitivity and specificity of the techniques used to identify the bacteria.
Known pathogens, including Aeromonas hydrophila, Aeromonas sobria, Edwardsiella tarda, and Streptococcus spp. were frequently isolated from diseased fish. Among the disease symptoms observed were skin ulcerations, exophthalmos, hemorrhages, dropsy and fin rot. The mortality rates associated to these bacteria were high and all cases were from farmed fish. Other authors reported similar findings with Aeromonas spp. (Sreedharan et al. 2012) and Edwardsiella tarda (Joh et al. 2013). These bacteria are known to be highly pathogenic in fish. Though 100% of the diseased fish reported in this study were from farmed fish, several cases of massive death of wild fish have been reported in Uganda without clear understanding of their aetiologies. Such mortalities could have been due to various diseases including those caused by bacteria isolated in this study.
The majority of the ‘other’ bacteria isolated have been associated with a few cases of disease in fish elsewhere with varying pathogenicity. Such cases include; Chryseobacterium indoligenes (Bernardet et al. 2005; Pridgeon et al. 2013), Plesiomonas shigelloides (Cruz et al. 1986; Nisha et al. 2014), Pseudomonas aeruginosa (Thomas et al. 2014), Citrobacter spp. (Jeremic et al. 2003), Serratia marcescens (Baya et al. 1992), Klebsiella spp. (Daskalov et al. 1998), Pseudomonas alcaligenes (Xu et al. 2015) and Pseudomonas fluorescens (Csaba et al.1984). It was difficult to associate these bacteria with disease in this study, since all disease cases with their involvement occurred as concurrent infections with known pathogens. In addition, they were commonly isolated from apparently healthy fish. No information was available to us about the pathogenicity of Comamonas testosteroni, Pseudomonas stutzeri and Ralstonia picketti in fish. Even in this study, they were only isolated from apparently healthy fish. Many fish pathogenic bacteria although ubiquitous in the aquatic environment, it is dependent on stress in the fish host to cause disease (Lio-Po and Lim 2014; Barton 1997; Walters and Plumb 1980). The increasing intensification of fish farming, a practice characterized by high stocking density, low water quality and increased human interference; together with the current trend of environmental deterioration may make these rare bacteria more important fish pathogens in the near future. This is supported by the fact that the majority, including Plesiomomas shigelloides (Jönsson et al. 1997), Chryseobacterium spp. (Douvoyiannis et al. 2010), Comamonas testosteroni (Tsui et al. 2011), Ralstonia picketti (Makaritsis et al. 2009) and Pseudomonas stutzeri (Noble and Overman 1994) have caused disease in humans.
Ponds had a significantly higher prevalence of bacteria (p < 0.05) as compared to cages and the wild water. In addition, though the diversity was low in tanks, the 2 tanks examined were both positive for Aeromonas hydrophila. This could be attributed the high density of fish, low water quality, increased human activity, water re-circulation, stock movement and high organic matter, characteristics of aquaculture systems especially earthen ponds. The high density of fish in aquaculture facilitates infection and transmission of bacteria (Penders and Stobberingh 2008). Direct contamination of pond waters by bacteria from surrounding soils could also contribute to the observed high prevalence and diversity of bacteria in earthen ponds. Tanks having the lowest diversity could be due to their more closed system which cannot be freely accessed by bacteria from the surrounding soils and runoff water.
Some bacteria, all Gram-negative rods, were not properly identified. These were characterized by inconsistent biochemical reactions at conventional biochemical tests and unacceptable profiles with API 20NE and API20E test kits. Failure to identify these bacteria could have been due to gaps in the diagnostic ability of the techniques used or these bacteria could represent new genera/ species of bacteria infecting fish.
Both Aeromonas spp. and P. shigelloides showed 100% susceptibility to at least 10 of the 14 antibiotics tested. However, 100% resistance was observed against penicillin, oxacillin and ampicillin. The two genera of bacteria expressed varying levels of resistance to erythromycin. Such high susceptibility levels of fish bacteria have been reported elsewhere (Aravena-Román et al. 2012; DePaola et al. 1995; Gonzalez-Rey et al. 2004). In a study by Akinbowale and others (Akinbowale et al. 2006), the Plesiomonas isolate tested was sensitive to all drugs tested except kanamycin, however, their study observed varying levels of resistance by other bacteria to several antibiotics. The high levels of resistance to the β-lactam antibiotics have been reported in genus Aeromonas (Penders and Stobberingh 2008; Jalal et al. 2010) and several other Gram-negative bacteria. This kind of resistance has been attributed to their intrinsic resistance, often chromosomal mediated and transferable to successive progeny during cell division (Kümmerer 2009). Aeromonas spp. is among the few bacteria with several chromosomal β-lactamase genes, including cphA, cepH and ampH which encode class B, C and D β-lactamases respectively (Balsalobre et al. 2009; Balsalobre et al. 2010). However, susceptibility of Aeromonas spp. to ampicillin has been reported (Maniati et al. 2005). Extended-spectrum β-lactamases (ESBLs) were not produced by 90.9% of Aeromonas hydrophila in South Africa (Ramalivhana et al. 2010). Unlike this study, different authors have reported up to 100% resistance against various antibiotics in fish farms (Newaj-Fyzul et al. 2008; Sarter et al. 2007; Castro-Escarpulli et al. 2003). Multiple resistance has been reported in fish (Ribeiro et al. 2010; Akinbowale et al. 2006); and in the aquatic environment (Li et al. 2010; Odeyemi and Ahmad 2017). Our findings indicate very low levels of acquired antibiotic resistance, suggesting minimal use of antibiotics in aquaculture in Uganda. This is in agreement with the findings by Walakira and others who reported use of antibiotics in fish by a few hatchery operators (Walakira et al. 2014). Surprisingly, high levels of antibiotic resistance have been observed in the humans and livestock (Byarugaba 2004; Kalule et al. 2012) in the same area. This could indicate minimal transmission of AMR from terrestrial to aquatic environments. Besides, intensive aquaculture is in its infancy in the country, it might be just a matter of time for resistant bacteria to evolve.
The susceptibility patterns of Aeromonas spp. were very similar to that of P. shigelloides. These bacteria are closely related and can share resistance determinants. The bacteria were isolated from common environments and are therefore likely to be exposed to similar antibiotic resistance selection pressure. Besides, this finding may be in support of previous reports that genus Aeromonas is an indicator bacterium for antimicrobial resistance in the aquatic ecosystem (Baron et al. 2017; Penders and Stobberingh 2008; Usui et al. 2016).
Bacteria from farmed fish showed some resistant traits to streptomycin, erythromycin and tetracycline unlike bacteria from wild fish. Similar studies have reported high levels of AMR in bacteria isolated from farmed fish (Newaj-Fyzul et al. 2008). This could be due to some low levels of direct use of antibiotics on fish farms for treatment and/or growth promotion. It could also be due to transfer of resistant bacteria from humans and livestock to fish bacteria as previously reported (Van den Bogaard and Stobberingh 2000; Akinbowale et al. 2006). Use of animal wastes to fertilize fish ponds (Sørum and Sunde 2001; Shah et al. 2012) and the naturally occurring resistant bacteria in the aquatic environment and soils (Cantas et al. 2013) could contribute directly to this effect by passing over antibiotic resistance genes to fish bacteria. There is a high risk of infection and spread of bacteria when animals are living in high densities like in intensive aquaculture (Penders and Stobberingh 2008).
Conclusions
This study identified the different bacteria infecting African catfish and Nile tilapia and determined the antibiotic susceptibility of selected fish bacteria in Uganda. This information is vital for fish disease management in the country and provides baseline data for future reference. Major fish bacterial pathogens are prevalent in Uganda and could partly be responsible for the observed reduction in fish production in the country. Fish bacteria in Uganda is highly susceptible to a number of antibiotics, however, accelerated emergence and spread of AMR as a result of the increasing intensification of fish farming, and the introduction of resistant bacteria from the terrestrial environment is a likely scenario in the near future. Efforts should be in place to control fish bacterial diseases and, emergence and acceleration of antibiotic resistance in fish bacteria in Uganda. Focus on alternative control strategies for bacterial infections in farmed fish without the use of antibiotic should be encouraged. To be able to control acute cases of bacterial infection in fish farming in Uganda without being hampered by antibiotic resistance, and to minimize the risk of transferring antibiotic-resistant bacteria to the human population through fish products, use of antibiotics in fish farming in Uganda should be discouraged. Characterization of the isolated bacteria including pathogenicity studies is urgently needed so as to clearly understand their implication on fish and human health in the country. Antibiotic susceptibility should be determined for individual bacterial species in wider studies. In addition, future studies should focus on use of more reliable bacteria identification methods such as sequencing of the 16S rRNA gene to clearly elucidate the situation.