Introduction
The geographical location of Indonesia’s islands on the planet connects two oceans, i.e., the Pacific and the Indian Ocean, affecting Indonesia’s waters as a place of water masses interchange with different characteristics. The Indian Ocean is warmer than the Pacific Ocean. Temperatures vary according to location and the ocean’s currents. At the same time, the water exchange occurs in the middle of the Indian Ocean and the Pacific Ocean, which causes the Indonesian Sea to be a quality habitat for various marine fishes, estimating more than 33,000 fish species alive. About 70% of them inhabit the Pacific Sea waters, which exists as the Great Sea, and more than 20% of the rest come from the Atlantic Ocean, while 8% come from the Indian Ocean with warmer waters characteristics (Vishnupriya et al., 2022). Several 4,954 species in 324 families are from the area currently known (Peristiwady, 2021). Next, over half of the total number of specimens collected present the families of Pomacentridae, Labridae, Gobiidae, Apogonidae, Chaetodontidae, Serranidae, Acanthuridae, Lutjanidae, Carangidae, and Scaridae. These families comprise about 48.9% of the total species reported. The Ichthyofauna dominates reef-associated fishes and pelagic or benthic fishes inhabiting offshore habitats.
Lombok Island, located in eastern Bali Island, links the Java Sea to the Indian Ocean and is well known as one of the dominant main water flow passages in the Indonesian Sea. Along the south coast of the Sumatra-Java and southern waters of the Lombok Island causes an increase in water mass (upwelling) in the east season (June to November), which causes the increasing inside seawater mass with low-temperature characteristics, high salinity, and rich in nutrients. This phenomenon increases the amount of chlorophyll and water fertility and then causes fish to be abundant (Eisele et al., 2021; Hendrawan & Asai, 2011).
The Genera Sardines (Sardinella) from the family Clupeidae, which come from the Mediterranean Sea, are small pelagic tropical marine fish having the principal economic value (Hunnam, 2021). Sardines, in Indonesia waters, are the third largest leading commodity after tuna and shrimp, whose producing shaws tend to increase and have been hazardous due to over-exploitation, pollution, habitat destruction, and climate change in the last ten years (Suherman et al., 2020). The other small pelagic fish, Spratelloides delicatulus, is one type of anchovy in the waters south of Lombok and likewise has experienced exploitation (Mahrus et al., 2022).
The large Sardine area in Indonesia is the Bali Strait, which is close to the Lombok strait, dominated by Sardinella lemuru Bleeker 1853 (Tinungki & Sirajang, 2019). They consist of five genera, and at least 21 unique species are ambiguous because of many similarities that are difficult to distinguish between one species and another (Thomas et al., 2014). Sardine species in Indonesia have names. i.e., Clupea (Harengula) longiceps (C.V.), S. lemuru Bleeker, 1853, Sardinella Amblygaster, Sardinella atricauda, Sardinella longiceps, Sardinella fimbriata, S. clupeoides, S. lemuru (Bali Sardinella), etc., and all species in Indonesia are called lemuru (Hunnam, 2021; Suherman et al., 2020).
The preliminary studies reported that the genetic diversity of Sardine is slightly higher than other marine fishes even though there is low genetic distance and variation between their populations, such as S. lemuru Bleeker 1853 in the Bali Strait, northern and southern-east Java (Sartimbul et al., 2018; Thomas et al., 2014). The use of mitochondrial DNA c oxidase subunit 1 (CO1) genes is a very effective method for identifying characterizing and identifying animal species quickly and accurately (Mohanty et al., 2015; Wang et al., 2018). The effectiveness of the CO1 genes is low in intraspecific variation, while interspecific variation is high, especially in adjacent taxa (Choi et al., 2020). The Fauna of Indonesia already has DNA barcodes of a few numbers on the report Center for Indonesian Institute of Sciences, consisting of mammals, birds, Komodo dragons, and insect pests (Rahayu & Jannah, 2019).
Bingpeng et al. (2018) said that the molecular technique is an alternative method for difficult-to-identify species. The number and names of Sardine species in Lombok, based on molecular characters using DNA CO1 genes or other genes until present, have not yet been reported, so it is still ambiguous taxonomy. Based on taxonomic uncertainty, this research aims to determine molecular characteristics, genetic diversity, and kinship for Sardines in Tanjung Luar fish landing center using the sequence of CO1 genes. It is located in Tanjung Luar Village, East Lombok Regency, West Nusa Tenggara Province, and has an area of approximately 222 km2 with a coastline of 220 km and a sea area of 1,074.33 km2. It has significant opportunities to develop fishery activities for capture fishing and culture fishery. To utilize fish resources optimally, wise management of marine and fisheries resources is needed. Therefore, this research will be crucial in elucidating.
Materials and Methods
The Sardine sampling lasted from May 10 to September 12, 2024, in the fish landing center of Tanjung Luar, caught from several locations on Lombok Island by small-scale fishermen (Fig. 1). It is located in Tanjung Luar Village, East Lombok Regency, West Nusa Tenggara Province. The distance between Tanjung Luar Village and the capital of East Lombok Regency is 18 km, and 65 km to West Nusa Tenggara Province. Tanjung Luar Village has an area of approximately 222 km2 with a coastline of 220 km and a sea area of 1,074.33 km2. It has quite a great potential for developing fisheries businesses for both capture fisheries and aquaculture.
Collecting Sardines samples amounted to 100 individuals using random methods. A Sardine grouping to the genus Sardinella based on their morphological characteristics, including body shape, body size, head shape, head size, and others (Carneiro et al., 2019). Sardine samples used for molecular genetic analysis were mantle tissue (±1 cm) cut using surgical scissors. Next, put and preserve it into 5 mL sample tubes with 96% alcohol and save it at the Immunology Laboratory, Faculty of Mathematics and Natural Sciences, University of Mataram.
Taxonomic identification was carried out up to the species level whenever possible. Genetic diagnosis, i.e., meristic counts and proportional measurements of collected specimens, was accomplished following (Sidiq et al., 2021). The morphological characters were observed and recorded in every sample in the premade datasheet. Determining fish species used nomenclature following the Food and Agriculture Organization (FAO) Fish Identification Sheets (Hendrikx, 2014).
There are 20 individuals for molecular characteristics identification. Fixating them previously first used 96% alcohol and saved in the Immunology Laboratory, Sciences Faculty, University of Mataram. Molecular identification in this study used the CO1 genes. They are the best solution currently identifying several fish and other fauna species because they work quickly and accurately (Bingpeng et al., 2018). Additionally, these genes can recognize species of conservative nucleotide base sequence and undergo only minor variations, deletions, and insertions (Miya et al., 2015; Phillips et al., 2019).
The Sardine sample used in extraction and DNA amplification of the DNA Sardines amounted to 20 individuals from 100 Sardine samples. Extraction and amplification of Sardine DNA has the potential as a candidate DNA marker for Sardine species using a commercial kit (DNeasy Blood & Tissue Kit, Jena Bioscience, Jena, Germany) with several modifications according to tissue type or using 10% Chelex solution (Barrientos-Villalobos & Schmitter-Soto, 2019) at 95°C. DNA was extracted from a small piece of ethanol-preserved muscle tissue using a slightly modified method according to the standard DNA method for fish (Hellberg et al., 2014). The amplification of CO1 rRNA target genes utilized the universal primer pair, ie. Primer Forward 5’-ATCTTTGGTGCATGAGCAGGAATAGT-3’, and followed the Primer Rivers: 5’-ACTTCAGGGTGACCGAAGAATCAGAA-3’ (Nuryanto et al., 2019; Ward et al., 2005).
In this study, the amplicon is approximately 680 bp from the 5’ region of the mitochondrial COI gene. Polymerase chain reaction (PCR) amplification conditions are as follows: denaturation for 5 min in 35 cycles at temperatures of 95°C (30 s), 50°C (30 s), and 72°C (50 s). The PCR reaction used a total volume of 50 μL containing one μL of template DNA, 10 mm Tris-HCl (pH 9), 50 mm KCl, two mm MgCl2, 0.2 μm of each primer, and 0.2 mm of each dNTP, and one U Taq polymerase. PCR products migrated using a 1.2% agarose gel visualized under UV light and documented using gel photographs. Then, PCR products through PT Genetica Science Indonesia for sequencing. The results of DNA sequencing of the CO1 genes were sent to researchers in file AB1 through email. Next, conduct a match between the electropherogram and the DNA sequence obtained in this study. The sequences obtained from a pair of CO1 primers were used as forward and reverse primers, each with reverse complementation to be the reverse. The next step is to align the two sequences using the W cluster menu (Katoh et al., 2019).
The CO1 gene sequence data was analyzed using MEGA 11 Software (https://www.megasoftware.net/), edited, and aligned using Clustal W to see the diversity of nucleotide bases (Tamura et al., 2021). The analyses of sequences used references of various species belonging to the Clupeidae family by the National Center for Biotechnology Information (NCBI) GenBank. Sequence alignments are also subjected to a basic local alignment search tool (BLAST) nucleotide search to determine their identity.
CO1 gene sequence data for all Sardine species available in GenBank were included in subsequent analyses using MEGA 11 Software (Tamura et al., 2021). This study elucidates the consensus sequences in each specimen using the Staden Package. Next, sequence differences within and between species were analyzed and tested using bootstrap tests with 1,000 replications.
Median-joining haplotype networks of both COI gene sequences (688 bp) used DnaSP v6 for specimens of Sardine (Rozas et al., 2017). The use of analysis of molecular variance (AMOVA) to test genetic differentiation among several groups based on molecular characteristics (Doğan & Doğan, 2016).
COI gene sequences were aligned, and the optimal substitution model used MEGA11 (Tamura et al., 2021). Phylogenetic trees were constructed based on the concatenated data of the two gene sequences using neighbor-joining (NJ) tree methods with 1,000 bootstrap replicates. Genetic distance matrix calculations used the Kimura-2 parameter model implemented in pairwise distance calculation in MEGA 11 Software.
Results
Sardines are a species of Actinopterygii fish from the Clupeidae family. The morphological form of the Sardine (Fig. 2) has characterization, i.e., its slender or elliptical body with a greenish-blue color on its back and a silvery underside. The body length can reach up to 23 cm, with an average length of 20 cm. The abdomen is slightly rounded, and the spines are blunt and not prominent. From the top of the gill cover to the base of the tail, there is a row of 10–20 black dots. The fins are yellowish-gray, but the tail fin is blackish, and the tip of the snout is also blackish.

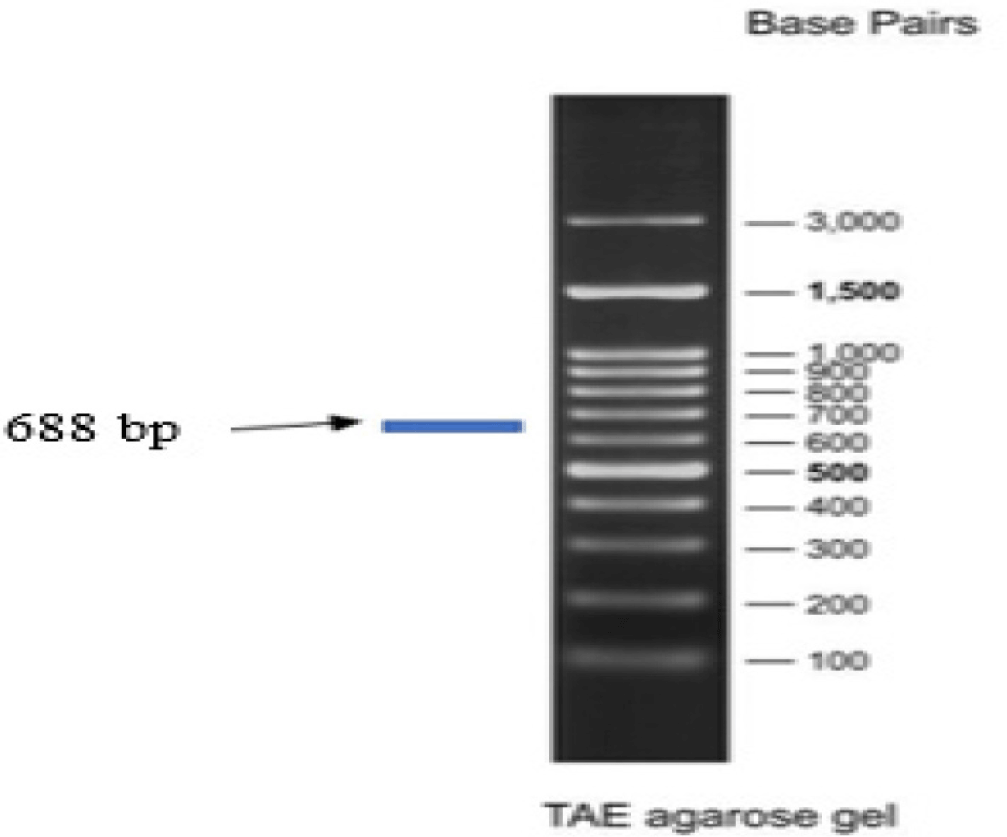
DNA sample isolated from the tissue of Sardine using a commercial kit (DNeasy Blood & Tissue Kit, Qiagen Cat. No. 69504, Jena Bioscience) successfully indicated by the qualitative test results of 1% agarose gel electrophoresis. The amplification of COI gene fragments from Sardines using a pair of universal COI primers, LCO-1490 and HCO-2198, successfully got amplicon lengths of around 688 bp (Fig. 3).
The results of tracking the CO1 gene sequence for 13 Sardine species in the NCBI and barcode of life data systems (BOLD) databases show that not all the BLAST sequences have the same length as the sample sequences (Table 1). BLAST aims to find regions of local similarity between sequences.
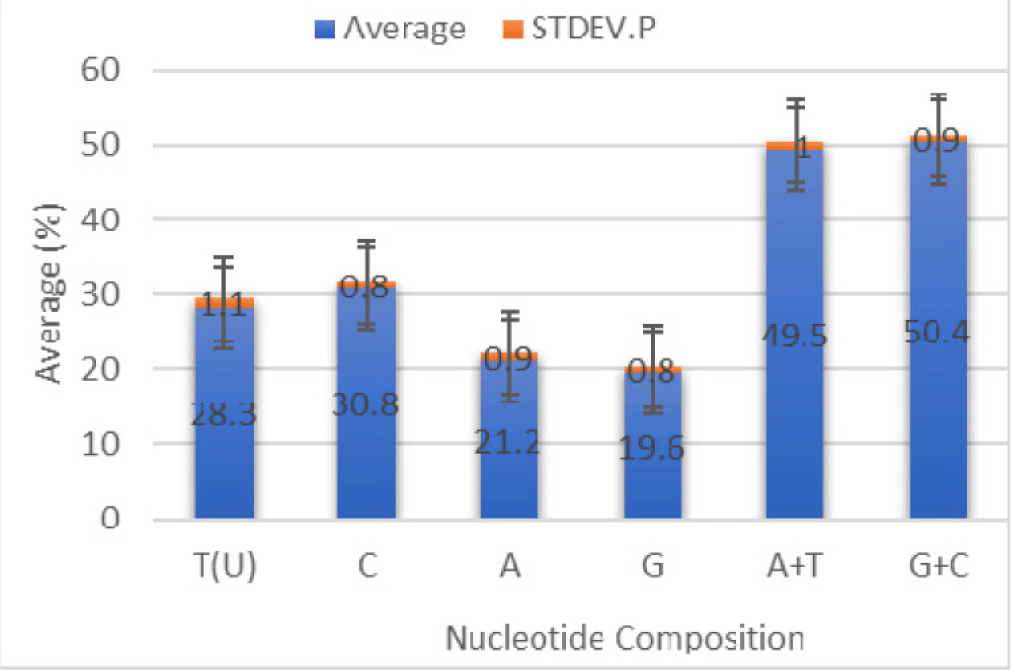
The nucleotide composition found in the COI fragment of Sardine from Lombok Strait is T = 29.0%, C = 31.1%, A (20.9%), and G (19.0%), with the nucleotide composition A + T = 49.9% and G + C= 50.1% (Table 2). Fragment length of the COI of Sardines ranged from 436 to 688 bp.
Data for comparison of nucleotide composition of 14 individuals of Sardine based on CO1 gene sequences is presented in Fig. 4 and is homogeneous.
The genetic distance ranges from 0.00 to 0.33 in CO1 fragments of 14 individual Sardines (Table 3).
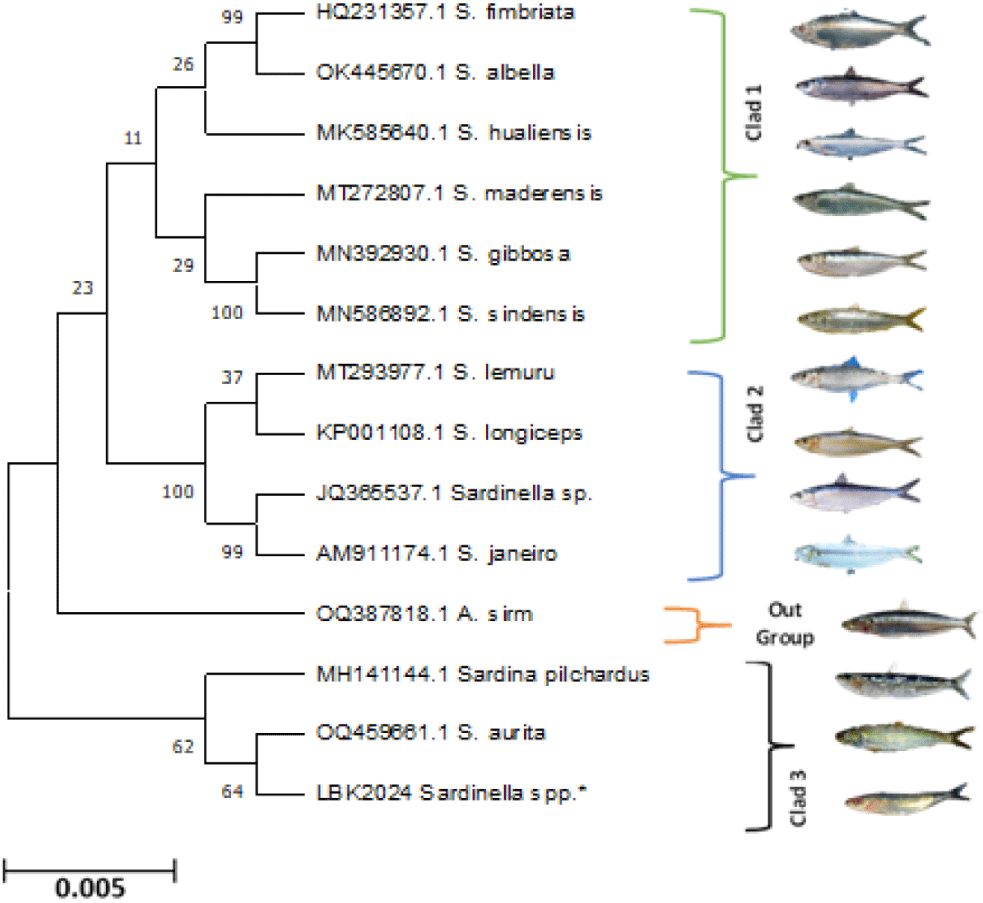
The maximum likelihood (ML) tree revealed distinct clades separated based on the genus of Sardine, Family Clupeidae (Fig. 5).
Discussion
The morphological characteristics of Sardines are generally found at fish landing center-tanjung luar-east Lombok, similar to Sardines in Indonesia waters, and often consist of S. lemuru, S. atricauda, S. longiceps, S. fimbriata, S. clupeoides, and S. leiogaster. Aside from the Sardine names, other names of Sardines from different genera are Amblygaster sirm, Herklotsichthys quadrimaculatus, Herklotsichthys dispilonotus, and Escualosa thoracata (Hunnam, 2021).
The qualitative Mitochondrial DNA isolation test in Fig. 3 showed a thin band on the agarose gel 1%. This success is similar to the isolation results in the Mitochondrial DNA of marine fish tissues, as reported in previous studies by Mascolo et al. (2019). Amplicons of 688 bp can be polymorphisms causes the possibility of intraspecies occurrence can be polymorphisms because of the possibility of intraspecies occurrence using universal primers (Yang et al., 2014). The CO1 genes have the advantage that their universal primers are very robust, so they can recognize the 5’ end of most animal groups, although not all, but represent most of the phylum Animalia (Ward et al., 2005).
The results of sequence homology comparisons with the NCBI and BOLD databases show that Sardinella spp., research samples from Lombok, have sequences that are similar to the COI gene fragment of the Sardinella aurita. The level of similarity between the two species ranges from 95.00% to 99.00% (NCBI) and 95.0–100.00% in the BOLD system. Accuracy and reliability assessments of sequences generated from the BOLD structure outperformed GenBank in terms of performance for Sardinella spp. (100% and 99%), respectively. From this data, sequence homology results with the NCBI database and the BOLD system show that the research samples from Lombok have sequences that are similar to the COI gene fragment of the S. delicatulus species with very high similarity (99%). The DNA sequence of 688 base pairs encodes 229 amino acids, with the amplified regions located at nucleotide positions.
The sizeable A + T nucleotide composition in Table 2 is in Sardinella hualiensis (46.8%) for the CO1 DNA fragment. In contrast, the great G + C nucleotide composition (53.2%). The mean nucleotide composition among the sequences of 14 Sardinella spp. individuals were estimated as T = 28.3 ± 1.1%, C = 30.8 ± 0.8%, A = 21.2 ± 0.9%, and G = 19.6 ± 0.8%. The content means AT is 49.5 ± 1.0%, and GC is 50.4 ± 1.9%. Nucleotide composition can support fish identification because differences in nucleotide composition indicate variations in nucleotide sites. These varied sites become specific characteristics that can differentiate fish species. According to Kombong & Arisuryanti (2018), differences in the nucleotide composition of the mitochondrial COI indicate genetic variation.
Genomes of fish and amphibians are GC homogenous, whereas birds and mammals are AT/GC heterogeneous, and the exact reason for this phenomenon remains controversial. Data GC/AT on the CO1 gene fragment is homogeneous, as in Fig. 4. The results are supported and compatible with previous research on teleost fish genomes (Symonová & Suh, 2019).
The average percentage of primary composition of CO1 gene fragments in various Sardine species does not match research on Indian freshwater fish with an amplicon of 700 bp and the average nucleotide composition is A: 23.2%; T:29.3%, G:18.9%; C: 28.6%, while in this study A: 21.2, T: 28.3, G: 19.6, C: 30.8% (Lakra et al., 2016). Likewise, the results on Mugil cephalus with rich and poor control areas at G content = 17.5% (Jamandre et al., 2014).
The calculation of genetic distances using the Kimura-2-parameter method integrated into MEGA 11 is the matrix of distances between base pairs between sequences that approach evolutionary distances. The highest intergeneric divergence (0.33), as presented in Table 3, was between Sardinella spp. and S. fimbriata. In contrast, the highest intrageneric divergence (0.30) is available on Sardinella spp. and S. aurita. S. fimbriata generally showed high sequence divergence from other Sardine species (0.22–0.33) studied here. The lowest intergeneric sequence divergence was between S. hualiensis, Sardina pilchardus, and A. sirm (0.00). In contrast, the great size intrageneric distance is Sardinella spp. with S. fimbriata, S. lemuru, and S. aurita (0.31).
Genetic distance is a measure of the transformation divergence between copies of homologous genes which share a common ancestor (Doğan & Doğan, 2016). Nei’s genetic distance value ranges from 0.0005 to 0.1006. The genetic distance value of 0.010–0.099 is in the low category. The genetic distance analysis found that the lowest genetic distance (0.00) occurred in S. hualiensis and Sardina pilchardus. Maximum and minimum genetic distance was recorded as 0.33 to 0.00 in the Sardine family Clupeidae, called the high distance category. This study is similar to research reported that average heterozygosity ranged from 0.3009 to 0.3744 (Mahboob et al., 2019). This information on genetic polymorphism is helpful for the concerned authorities evolving strategies to conserve the diversity of tilapia in the country.
The analysis of the ML method using the Kimura-2 model showed the kinship between species based on the length of the branch lines. Different line lengths indicate the level of evolution of each species. Longer lines indicate further evolutionary distances, while shorter lines indicate closer evolutionary distances of a species. All Sardine species in this phylogenetic tree form clades and one individual (A. sirm) is outside the group (Fig. 5), while in the NJ, A. sirm is inside the group. In contrast, the unweighted pair group method with arithmetic places A. sirm and Sardina pilchardus outside the group, while the ML method places A. sirm alone outside the Sardine group, the estimation methods of ML is the best model because it has the minimum evolutionary change compared to the UPGAMA, dan NJ methods.
The results of nucleotide alignment along the 688 bp mitochondrial DNA COI gene fragment support this identification result, as presented in Fig. 5. Most of these sequences matched to reference sequences on GenBank or BOLD databases with more than 95% identity. The overall mean distance of the Sardine sequences was 23%. Analysis based on the phylogenetic tree shows that all Sardine members of the family Clupeidae are monophyletic and grouped into three Clads, namely Clad 1 consists of S. fimbriata, Sardinella albella, Sardinella hualensis, Sardinella maderensis, S. gibbose, and Sardinella sindensis; Clad 2: S. lemuru, S. longiceps, Sardinella janeiro, and Sardinella sp.; Clad 3: Sardina pilchardus, S. aurita, and Sardinella spp*., while A. sirm is out of the group. The reconstruction of the phylogenetic tree in Fig. 5 confirms that Sardines from the fish landing center of Tanjung Luar-East Lombok and S. aurita are in the same clade as S. aurita and have a very close relationship. The same results in preliminary research in Lombok Strait, but using a different gene fragment, the 12S gene, found that Sardine (S. lemuru) and S. aurita have a very close relationship (Mahrus et al., 2012).
A phonetic ML tree using K2P distances illustrated CO1-based genetic divergence among intra and inter-specific hierarchical units. There is grouping on All species. Thus, the COI gene correctly identified all species. Bootstrap analysis consensus trees with 10,000 replicates obtained from ML and MP showed similar patterns in the genetic relationship among the Sardine species studied. There was no haplotype sharing or overlapping. An average haplotype diversity (Hd) using DNAsp average of 0.9743. They mean the genetic diversity of Sardine is high. However, the population structure is low (Fst = 0.37488). The phylogenetic trees using the haplotypes for COI genes could unambiguously differentiate all the 14 species of wild in marine waters. Population structure plays the role of analyzing gene expression in the population (Fachrul et al., 2023). The analysis of molecular data is needed to recognize the extensive phylogenetic relation, investigate population composition inside a species, and resolve taxonomic ambiguities, especially in Sardine species, and has proved to be principal (Devanand et al., 2020). It is suggested that CO1 genes, as DNA barcoding, was a build-on tool to taxonomists, besides conventional taxonomy, could help to find a solution to the taxonomic ambiguities of Sardine taxonomy especially.
The results of this study, which has a low population structure with high genetic diversity, are similar to the results of research on an economically important fish species (Lutjanus jocu) reported by Verba et al. (2023). Preliminary research also reported similar cases of low population structure and genetic diversity in Rhinolophus blasii in Europe (Jakab et al., 2021). Factors that influence low population structure consist of the availability of low amounts of food, predators, competition with living creatures of the same species or other species, climate, changes in habitat, and disease (Verba et al., 2023). Next, differences in depth and color in these fish are not significant enough to be found as an indication of new species. In addition, the distribution of larvae, juvenile movement, and migration of adult fish that are actively moving can cause a species or population in different areas to have genetic connections or connectivity.
Using molecular DNA barcoding can identify organisms based on short and explicit DNA sequences, one of which is the CO1 gene (Ward et al., 2005). Mitochondrial DNA of the CO1 gene is one of the markers to determine the population. Mitochondrial DNA markers can determine molecular diversity in animals because they are easy to explain and produce population connectivity in a short time. Population connectivity also affects the adaptation of a species. The information related to population connectivity can be a guide in carrying out conservation and recovery efforts of a population because it has a direct or indirect impact on a population’s adaptation to its environment. Verba et al. (2023) reported that low population structure is affected by factors as well as restricted nourishment availableness, hunters, rivalry with other species or taxonomic groups, climate, natural environment changes, and illness. Unfortunately, the factors have not been investigated yet in the present, so they will be a particular concern in the future. Finally, it hopefully supports management activities, determination of conservation areas, and sustainable strategies to maintain and preserve Sardine fish resources in Indonesia.
COI gene fragments successfully amplified Sardines with an amplicon length of around 688 bp from fish landing center-Tanjung Luar, East Lombok. The study indicates that the mitochondrial DNA COI gene sequence can identify wild Sardine species in high levels of genetic divergence (0.00–0.34). Data on Hd using DNAsp: 0.97436 means the genetic diversity of Sardine is high. However, the population structure is low (Fst = 0.37488). Besides that, the COI gene sequence can be acceptable as an authentic identification marker for various marine fish species.
