Introduction
Phylum arthropoda has 50,000 recorded species and been reported to contribute 80% of other phyla (Poupin & Juncker, 2010) Brachyura crabs with the rocky coastal habitat in the Indo-Pacific region have a quite high species number. This is due to the Indo-Pacific and Indo-Malayan regions are part of the tropical region with high mangrove forest covers so the diversity of Brachyura lives in this area is relatively high (Castro et al., 2015).
Crabs are ten legs animals that belong to the order Decapoda and Brachyura infraorder (Poore, 2004). The morphology of crab body generally consists head, chest and abdomen. The head and chest are enclosed by a carapace while the abdomen is bent under the chest (Yeo et al., 2008). Poore (2004) affirmed that identifying crab could be done through describing the shape, size and color of the carapace. Diversity in shape, color and size in infraorder of Brachyura frequently results in inaccuracy in morphological analysis to determine the species name. Tindi et al. (2017) stated that morphological identification techniques are usually hampered by the identification stages due to the phenomenon of cryptic species. Hebert et al. (2004) stated the limitation of morphology-based identification systems and the decline in taxonomic number indicated the need for new methods in socializing taxa. Thus, molecular identification using DNA barcode is required to minimize such error.
According to Tallei et al. (2017), DNA barcoding has been widely used by researchers to find out the genetic distance between species using protein-coding genes in mitochondrial DNA, that is cytochrome c oxidase subunit I (COI) gene. The advantage of DNA barcoding is that it can identify species that are difficult to differentiate morphologically (Hebert et al., 2004). In addition, DNA barcoding provides rapid and accurateness in species identification (Karim et al., 2016; Muchlisin et al., 2013). DNA barcoding technique is a system designed to carry out species identification rapidly and accurately based on the nucleotide base sequence of a standardized short marker gene, namely the COI gene. COI gene in the mitochondrial genome is the starting point in taxonomic and molecular phylogenetic research (Borges et al., 2016; Kress & Erikson, 2012). The COI gene becomes a species-specific marker for molecular identification, especially in animals (Rahayu & Jannah, 2019).
Several studies in the field of molecular DNA have been conducted in Manado Bay, North Sulawesi, Indonesia. Using the COI gene, Tindi et al. (2017) found Atrina vexillum had the highest percentage in the Bivalvia class, Genus Atrina. Furthermore, Triandiza & Maddupa (2018) examined whether the use of DNA barcode analysis with COI gene in Anomura crab was capable to identify the crab up to infraorder level.
Research in the molecular field has been carried out on several crab species of Brachyura Infraorder discovered in intertidal habitats in Manado Bay. Paransa et al. (2019) found the crab caught from the coast of Tanawangko Village, Tombariri District, Minahasa Regency, North Sulawesi, Indonesia has a circular convex carapace with a blackish green longitudinal striped motif, and having long greenish white stripes. Through molecular analysis, it was identified as Grapsus albolineatus Latreille in Milbert (1812) (Paransa et al., 2019). Furthermore, Siahaan et al. (2022) found a crab from the coast of Minanga, Manado City, North Sulawesi and identified as Ategratis floridus. This crab has an oval and convex carapace and has symmetrical stripes on the right and left sides with a yellowish green color. The comparison of current identified species with the two previous finding was presented in Table 1.
| Species | Carapace shape | Color | Source |
|---|---|---|---|
| Metopograpsus oceanicus |
The dorsal carapace is trapezoidal in shape narrowing at the bottom of the posterior. The margin of posterolateral and anterolateral parts has straight edges without teeth/spines. |
- A pair of claws which are shorter than the pereiopods, purplish-orange in color, have a rough white dotted motif and the fixed finger (polex) is white. - The carpus, propodus and dactylus segments have yellow setae with fine hairs. A pair of pereiopod at the merus part is black with brownish yellow motif. |
(Paransa et al., 2023) (current finding) |
| Grapsus albolineatus |
Carapace is circular. Convex-shape, has four lobes. |
There are linear lines with orange dots at the dorsal of carapace. | Paransa et al. (2019) |
| Ategratis floridus | Crab has an oval and convex carapace. | Carapace has symmetrical stripes on the right and left sides with a yellowish green color. | Siahaan et al. (2022) |
Numerous crabs are discovered living on the coast of Manado City. Through morphological identification, Amin et al. (2021) and Lepa et al. (2022) found two crab species living on the sandy beach, Ocypode ceratophtalmus and Ocypode kuhlii. Other species were also successfully identified by several researchers, including Eriphia sebana, A. floridus, G. albolineatus, Pilumnus vespertilio, and Metograpsus sp. (Lepa et al., 2022; Manik et al., 2020; Rustikasari et al., 2021). In this current research, it was found Metopograpsus sp. at the coast of Tambala, Tombariri District, Minahasa Regency. Genus Metopograpsus have six species and known as tidal crabs because they live on sheltered rocky beaches and mangrove forests. These species of Genus Metopograpsus have similarities in morphological characteristics making it difficult to be differentiated and identified morphologically. The objective of this research was to identify the morphology and molecular profile of crab caught from the coast of Tambala Village, Tombariri District, Minahasa Regency, North Sulawesi.
Materials and Methods
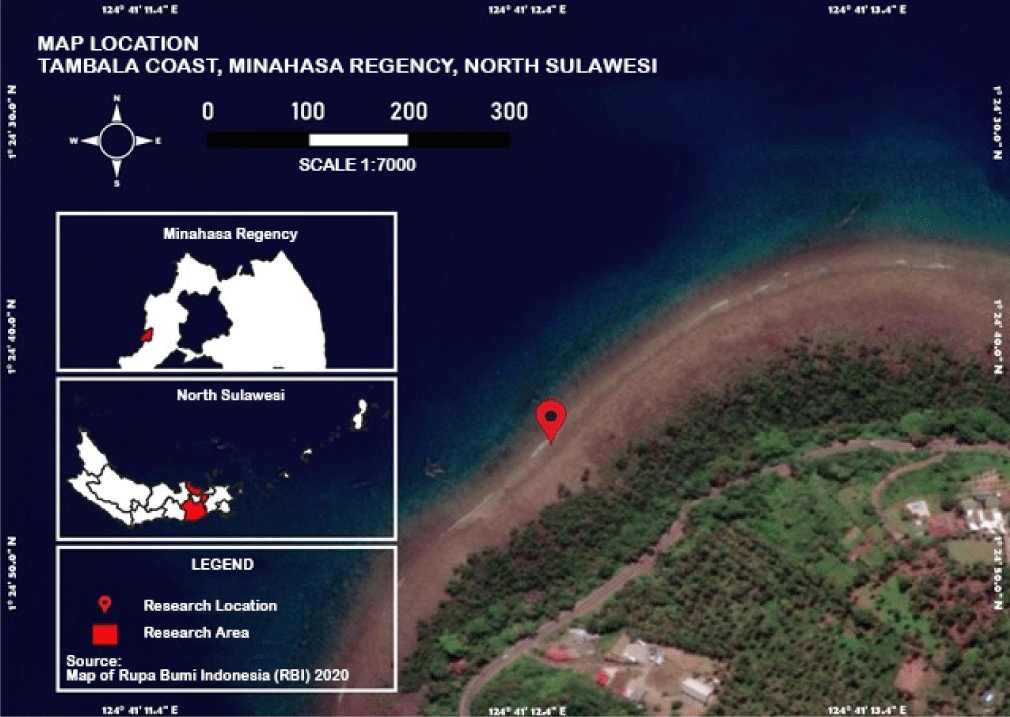
Research location was Tambala Coast, Tombariri District, Minahasa Regency, North Sulawesi at coordinates of 1°24’30.0”N and 124°41’11.4”E (Fig. 1). In specific, the location is rocky mixed with dead coral fragments, covered by mud and surrounded by mangroves Rhizophora mucronata. Rocky coastal is a habitat that is overgrown abundantly by filamentous algae (Kennish & Williams, 1997; Paransa et al., 2019). This habitat is also an ideal place for crab to look for food.
Crab samples were collected for three nights from the intertidal zone at night during the lowest tide. According to Lalli & Parson (2006), crabs are nocturnal organisms that actively search for food at night. The crabs were collected through exploring this intertidal area as far as 100 m, referring to the research results of Paransa et al. (2019) and Siahaan et al. (2022). Samples were caught using gloved hands and a flash light. The number of specimens collected were 30 individuals with the length of 3.2 cm in average. Exploration method is a data collection method carried out by exploring locations to observe living organisms, including invertebrate organisms, ecosystem types and vegetation in the studied area (Salvanes et al., 2018). A report by Paulay (2007) showed that crabs from the Grapsidae family in rocky habitats tend to inhibit rocky coastal locations or on artificial rocks such as seawalls and breakwaters.
Morphological determination of crabs in the Brachyura Infraorder was performed based on the characteristics of color, size and shape of the carapace (Poore, 2004), anterolateral, posterolateral, eyes, antennae, setae/fine hairs, propodus, lobes and various claws (Castro et al., 2015; Naderloo, 2017). Molecular analysis was carried out through DNA barcoding analysis include DNA extraction, amplification and electrophoresis.
Results
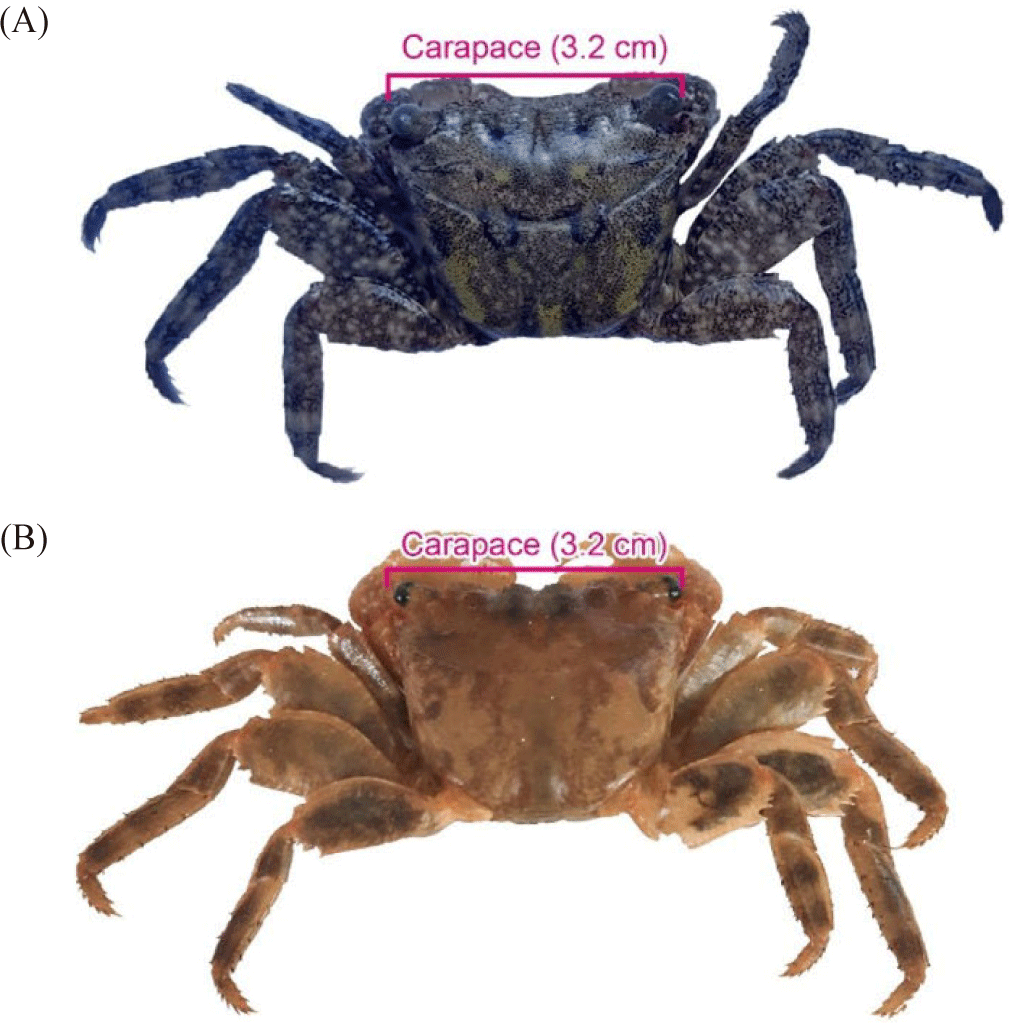
Brachyura crabs caught from the coastal area of Tambala Village, Tombariri District, North Sulawesi Province is displayed in Fig. 2. Morphologically, the Brachyura infraorder found in this research have a pair of claws which are shorter than the pereiopods, purplish-orange in color, have a rough white dotted motif and the fixed finger (polex) is white. The merus part found on both sides of its claws has crown-like spines located under the exorbital.
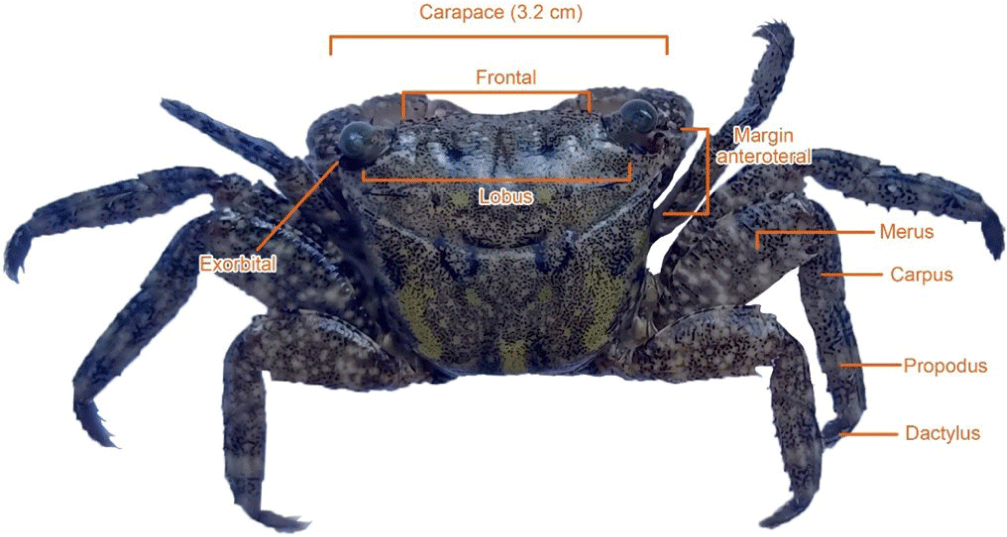
Pereiopods (walking legs) consist of the ischium, merus, carpus, propodus, and dactylus with different sizes. The carpus, propodus and dactylus segments have yellow setae with fine hairs. A pair of pereiopod at the merus part is black in color with brownish yellow motif.
The dorsal part of carapace is blackish green with light green and brownish spots. The dorsal carapace is trapezoidal in shape narrowing at the bottom of the posterior. The margin of posterolateral and anterolateral parts has straight edges without teeth/spines. Matching and differences in patterns and colors of the crab before and after soaking with alcohol is presented in Fig. 3.
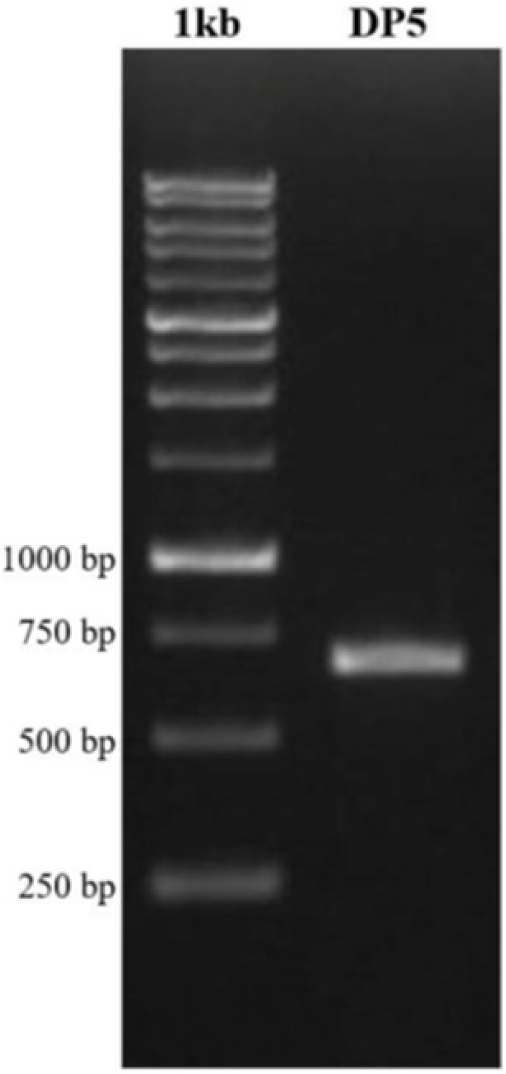
Extraction of genomic DNA was taken from pieces of muscle tissue specimen from Brachyura crabs. The result of extraction was an elution containing pure DNA mixed with buffer elution. The next step was amplification of the COI gene using primers designed by Folmer et al. (1994), namely F-LCO1490 with the pair of R-HCO2198. The primer pair used had successfully identified A. floridus crabs caught from rocky coastal habitats (Siahaan et al., 2022). The amplification results were followed by separation with 1% agarose gel electrophoresis. Next, the DNA of the polymerase chain reaction (PCR) product was visualized using a UV-Transilluminator and the success of the PCR was detected by the presence of a DNA band as shown in Fig. 4.
DNA sequencing was carried out by using the dideoxy method (Sanger Method). DNA editing was carried out using Geneious v5.6 software (Geneious®, Auckland, New Zealand) while the comparison using the MUSCLE algorithm (Kearse et al., 2012). Identification used the GenBank database (https://www.ncbi.nlm.nih.gov) with FASTA format DNA sequences, then referring to significant alignment with NCBI data using BLASTn (Zhang et al., 2000). Nucleotide sequence of the CO1 Gene (DP5) from muscle specimens of M. oceanicus is shown in Table 2.
Discussion
The Brachyura crab found in this research had a pair of claws which are shorter than the pereiopods, purplish-orange in color, have a rough white dotted motif and the fixed finger (polex) is white. This finding was in agreement with the research results of Amin et al. (2021) in the same genus, namely M. latifrons (White, 1847). The crab had a pair of claws of the same size with light brown color, the claw tips were brownish white with rough white spots.
Previous report by Naderloo (2017) explained that the merus part of the claws of the Metopograpsus genus had spines pointing out sharply on both sides. Furthermore, Manik et al. (2020) found that crabs caught from Manado Bay at Mokupa Village of District Tombariri identified as Metopograpsus sp. had a pair of purple claws with black spots and white fixed finger (polex). This finding was similar with the report by Naderloo (2017) where Metopogragsus messor and Metopogragsus thukuhar has purple claws. Claws function to hold and carry food, dig and as self-protection from enemies (Castro et al., 2015; Denny & Gaines, 2007). Morphological characteristics of several Brachyuran crabs were presented in Table 3.
| Species | Carapace shape | Color | Source |
|---|---|---|---|
| Metopograpsus oceanicus |
The dorsal carapace is trapezoidal in shape narrowing at the bottom of the posterior. The margin of posterolateral and anterolateral parts has straight edges without teeth/spines. |
- Claws which are shorter than the pereiopods are purplish-orange in color, have a rough white dotted motif and the fixed finger (polex) is white. - The carpus, propodus and dactylus segments have yellow setae with fine hairs. A pair of pereiopod at the merus part is black with brownish yellow motif. |
(Paransa et al., 2023) (Current finding) |
| Metopograpsus sp. | The dorsal carapace is trapezoidal. |
The dorsal of carapace is blackish green color. Claw is purple with black spots and white fixed finger. |
Manik et al. (2020) |
| Metopogragsus messor | The dorsal carapace is trapezoidal; Carapace with anterolateral margins distinctly converging posteriorly. | Claws are purple and tend to be lighter than M. oceanicus. | Naderloo (2017) |
| Metopogragsus thukuhar | The dorsal carapace is quadrate; Carapace with anterolateral margins slightly converging posteriorly, nearly straight. | Claws are purple. | Naderloo (2017) |
| Metopograpsus latifrons | The claws have light brown color, claw tips are brownish white with rough white spots; periopods are dark brown with irregular white spots. | Amin et al. (2021) | |
| Metopograpsus oceanicus | Distinctly trapezoidal, posteriorly narrowing carapace. | Mottled in black, brown, and light tan; Chelae are violet in life. | Paulay (2007) |
On M. latifrons (White, 1847), Amin et al. (2021) found four pairs of dark brown pereiopods with irregular white spots and there were fine hairs on the dactylus, propodus and carpus. Manik et al. (2020) also found that Metopograsus sp. had a blackish green color on the dorsal of carapace. A report by Paulay (2007) showed the carapace color of M. oceanicus is darker than that of M. messor. Carapace functions to protect the head and thorax with different color patterns in Grapsidae Family. The crab found had four lobes which were divided into two parts and a pair of eyes between the frontal parts. Paulay (2007) also found M. oceanicus and M. quadridentatus collected from the coast of Hawaii in the harbour area have similar anterolateral tooth behind the exorbital tooth.
A very significant difference between fresh and preserved crab can be seen in carapace color (Fig. 3). There are differences in the dactylus part of the Fig. 3A the setae/fine hairs are yellow, carapace still has many color patterns, including black, green, brownish and light brown. Meanwhile in Fig. 3B the concavity at the frontal part is no longer clearly visible. The leg part of merus, propodus and carpus paths appear black in the middle and brownish on the sides. On the merus at the first and second walking legs. The fine horizontal lines are clearly visible. At the end of the merus, there are spines at the bottom and top.
The DNA sample path was observed at the amplicon length of around 600–750 bp using a 10,000 bp DNA ladder primer LCO1490 and HC02198 (Fig. 4). The crab sample (DP5) was characterized by the appearance of a thick and clear band on the gel track. PCR product DNA was visualized using a UV-Transilluminator and PCR success was detected by the presence of a single DNA band of 750 bp (DP5). Based on the visualization results, it showed that the length of the DNA band was 500–750 bp (DP5).
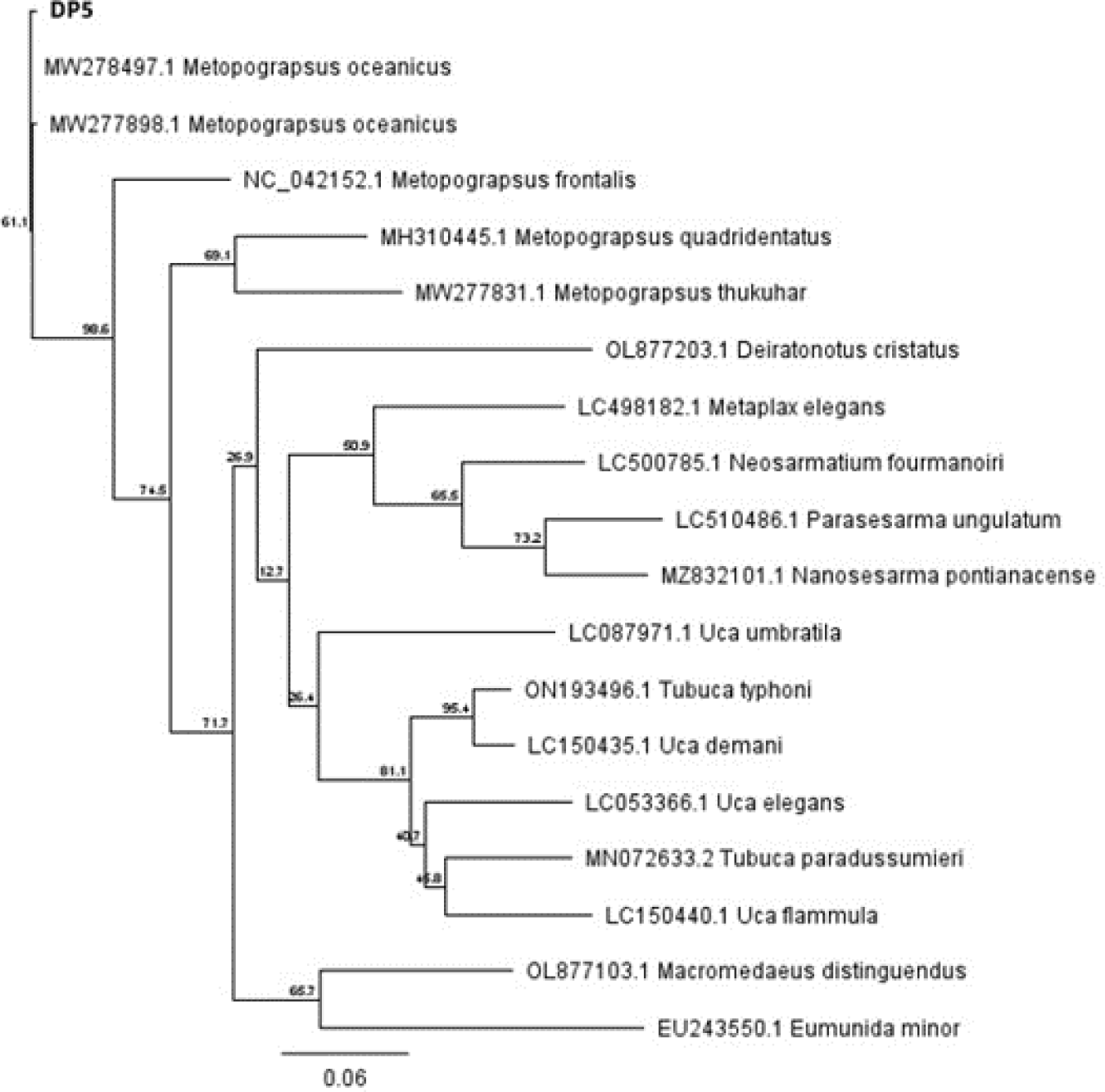
Based on GenBank from BLASTn, samples of crabs caught from the coast of Tambala Village, Tombariri district, Minahasa Regency used in this research was identified as M. oceanicus. This identification is presented in the form of a phylogenetic tree (Fig. 5). Construction of phylogenetic tree was carried out using the Maximum Likelihood Algorithm using PhyLUM v3.3.20180621 (Guindon et al., 2010) with Bootstrap 1,000 times, as shown in Fig. 6.
The Grapsidae Family has the closest relationship to sample DP5. The closest species to this sample is M. oceanicus from America. Sample DP5 also has relationship to M. frontalis, M. thukuhar, and M. quadridentatus. The following Table 4 presents the relationship level of M. oceanicus.
The level of relationship between the five Metopograpsus sp. (Table 4) can be seen from the Maximum Score and Query Cover which show a higher approach to the sample. According to Tweedie (1949), the five species of Metopograpsus have the same shape and size of carapace and celipids. Based on Table 4, the differences between the five species above are found in the color differences and carapace pattern. The interesting variations in color and pattern on carapace of Brachyura Order are caused by the presence of carotenoid pigments contained in the body tissue of the crabs. This is proven by pigment analysis research. Pigment analysis on the dorsal carapace of the M. oceanicus using Column Chromatography separation with developing solution of acetone and hexane (80:20) was identified two types of carotenoid pigments, namely β-carotene and echinenone.
Manik et al. (2020) and Adrian et al. (2021) found two species of Brachyura crabs on the coast of Tombariri District, specifically at Mokupa Beach. These crabs produced β-carotene, equinone, zeaxanthin, astaxanthin, and astacene pigments. Furthermore, Paransa et al. (2019) reported Grapsus sp. had linear lines with orange dots at the dorsal of carapace. This crab contains types of carotenoid pigments, such as β-carotene, equinone, canthaxanthin, astaxanthin and astacene. Mokoginta et al. (2021) reported the same species from the rocky coastal of Mangatasik Village, Tombariri District, Minahasa Regency, North Sulawesi Province produced β-carotene, zeaxanthin, lutein, β-kryptoxanthin, astaxanthin, astacen pigments. Research results of Paransa et al. (2023) showed that carotenoid pigments from G. albolineatus Latreile in Milbert (1812) identified using molecular docking and visualization with Autodock 4.2 and Discovery Studio/Biovia have potential as anti-aging.
The current research successfully identified M. oceanicus through DNA barcoding with COI gene. This species belongs to Brachyura infraorder. Brachyuran crabs play an important role in marine benthic communities, ranging from intertidal to deep waters (Boudreau & Worm, 2012). They prey for a wide range of invertebrates and vertebrates that are successful and versatile predators, preying at more than one trophic level. Crabs interact with the habitat and its inhabitants in a variety of ways, including providing habitat for smaller invertebrates and competing for food and shelter.
Using the same system, two species of Brachyura crabs were also identified, namely G. albolineatus, A. floridus (Paransa et al., 2019; Siahaan et al., 2022; Abbas et al., 2016) stated that Brachyuran crabs are one of the most diverse animal groups at the infra-order level. Thus, COI gene is very effective in identifying crab species. This is in line with statement of Costa et al. (2007) that the COI gene is very effective in identifying various decapod species. This method has been widely applied to identify new crab species. DNA barcoding can accurately identify and distinguish different species of crabs occupying an area.
The current study applies DNA barcoding and phylogenetic analysis accompanied by morphological analysis. The combination of these methods is essential for conservation purposes. However, data on the biology and ecology of the identified crabs is not yet available, such as feeding behaviour, food preferences, age of crab larvae, migration and so on. For conservation purposes, research on the biology and ecology of crabs is still very much needed.