Introduction
The diversity of life defines the biological world. Even maternal twins are not identical. Organisms have differences in one or more traits and it is known as the variability that causes genetic diversity. It can be assessed through the gene count from a specific pool of genes (Ahmad et al., 2022). Generally, genetic variation results from normal cellular processes or interactions of different organisms with the environment (Chauhan & Rajiv, 2010). Though, variations in environmental conditions and population density can cause considerable variability, genetic drift, migration, selection, and human interference contribute more significantly (Chauhan & Rajiv, 2010). These factors act continuously, causing changes in the population’s frequency of alleles as well as genetic diversity. The frequency of a few alleles is favored over others in domestication or artificial selection increasing selected alleles (Yilmaz & Boydak, 2006). Small-size populations have low genetic diversity and are also prone to genetic drift, which randomly reduces genetic variation (Lu et al., 2014).
Recently, the aquaculture sector is more focused on mitochondrial DNA (mtDNA) analysis for the assessment of deep divergence among populations or species. It is understandable because of its numerous benefits including easy collection and its inheritance to the progeny. In addition, due to the accumulation of divergence sequences, it is devoid of recombination and experiences a high rate of base substitution, which is becoming increasingly relevant in aquaculture for identifying profound divergences between populations or species (Mandal et al., 2012). Numerous researchers have reported that DNA barcodes that utilize a specific mitochondrial fragment, encompassing 600 base pairs recognized as cytochrome oxidase I (COI), are a quick, effective, and inexpensive method for gauging genetic diversity and kinship among species or populations (Hebert et al., 2004; Kamran et al., 2020). However, because of its sole maternal inheritance, a lot of researchers have expressed reservations regarding studies that rely entirely on mtDNA (Zhang & Hewitt, 2003).
Microsatellites are highly polymorphic, biparentally inherited, and found all over the euchromatic region of genomes. They have characteristics such as high variability, improved genomic coverage, high reproducibility, automation readiness, neutrality, and a lack of environmental fluctuations (Figueras et al., 2016). In aquaculture genetics, they are widely used for intrapopulation and interpopulation comparisons, identification of different strains, and genetic variability among individuals (Wattanadilokchatkun et al., 2022). However, due to the uncertainty of the ancestral information, cannot be used to deduce the genealogical patterns of relationships (Zhang & Hewitt, 2003). Thus, using both types of markers can help us to understand genetic diversity and relatedness among species or populations (Gariboldi et al., 2016).
Genetic diversity plays a vital role in providing the building blocks necessary for natural selection and facilitating the adaptation of organisms to new and challenging environments (Gandra et al., 2021). In the last 20 years, reports have shown a decrease in the genetic diversity of wild fish due to various factors such as lower recruitment, indiscriminate exploitation, overfishing, habitat destruction, migration route blockages, and human interference (Islam & Alam, 2004; Lu et al., 2014). Moreover, inbreeding, small population size, genetic drift, and restricted gene flow in captivity, subsequently result in the reduction of genetic variation among hatchery-raised fish has been reported by many investigators (Shah, 2004).
For the sustainability of aquaculture species and the conservation of wild stock, knowledge regarding the genetic structure of the stocks is a prerequisite (Sultana et al., 2015). However, information/literature on the genetic population structure of indigenous major carp in Pakistan, including Labeo rohita is scarce. L. rohita can be found in rivers of Pakistan, India, Bangladesh, and Burma (Dahanukar, 2010; Froese et al., 2013). It has high aquaculture and commercial importance worldwide, especially in Asia, and occupies the topmost rank among Indian major carp where it is consumed owing to its fame for being a tasty fish which results in its high market value (Rahman et al., 2005). However, over the past two decades, many investigators from Bangladesh, India, and Pakistan reported a decline in the genetic diversity of wild and captive stocks of L. rohita (Ahammad et al., 2022; Khan, 2008; Qadeer & Abbas, 2017). Therefore, the current study was designed to evaluate the genetic variation in both wild and captive populations of the indigenous culturable speciesL. rohita, by utilizing microsatellite markers and mitochondrial marker (CO1). Six fish hatcheries across Pakistan for captive-bred strains and two rivers, namely River Kabul and River Ravi for wild strains were selected to assess the genetic structure of their stock.
Materials and Methods
For the current study, six different strains of captive and two wild rohu (L. rohita) were collected from hatcheries and rivers across the country. The collection of strains was performed with the active collaboration of various collaborators at eight locations across Pakistan while local fishermen assisted in the collection of wild stocks from the Ravi and Kabul Rivers. The strains along with the study area given in Fig. 1 and for ease, the locations are also mentioned in the following abbreviated form.
-
USs (Upper Sindh Fish Hatchery, Province, Sindh; 25° 23’00” N 68°20’05” E)
-
SPs (Carp Hatchery and Training Center, Peshawar, Province, Khyber Pakhtunkhwa 34.0933° N, 71.5105° E)
-
WKs (Wild Strain from River Kabul; 33° 55′0″N 72°13′56″E)
-
WRs (Wild Strain from River Ravi; 31° 29’ 2.2272”N 74° 9’ 46.4292”E)
-
TMs (Tawakkal Fish Hatchery Muzaffargarh, Province Punjab; 30° 11’ 57”N 71°17’ 07” E)
-
MKs (Govt. Fish Hatchery, Mianchannu, Province Punjab; 30° 26’ 20.1732”N 72° 21’ 18.198”E)
-
CMs (Govt. Fish Hatchery, Charbanda, Mardan, Province Khyber Pakhtunkhwa; 34° 21’ 06” N 71° 58’ 22” E)
-
TKs (Tanda Govt. Fish Hatchery Kohat, Province Khyber Pakhtunkhwa33° 34’ 07”N 71° 24’ 01”E)
From all locations, twenty fish per population/strain were collected and their part of the caudal fins was removed, washed with 75% alcohol, and stored in 90% ethanol at a temperature of 4°C to enable future DNA extraction (Lutz et al., 2023).
A modified salt extraction method reported by Kumar et al. (2007) was employed to isolate the DNA from the caudal fin of each specimen. Briefly, the fin was cut into small pieces and dried on filter paper. Afterwards, 20 mg of macerated fin was placed in a 5 mL tube containing 1.5 mL lysis buffer (200 mM Tris-HCl (pH 8.0); 250 mM NaCl, 100 mM EDTA, 60 µL of 20% SDS, 10 µL Proteinase K (20 mg/mL). The contents were mixed well and incubated at 48°C for 3 hours in a water bath. Then, 800 µL of phenol-chloroform-isoamyl alcohol (25:24:1 ratio) was added to the lysed contents, and tubes were shaken manually for 5 min. The mixture was then centrifuged for 10 min at 10,000 rpm, and the supernatant was collected in a tube containing 800 µL of chloroform-isoamyl alcohol (24:1 ratio). The mixture was mixed well manually and again centrifuged, and the supernatant was collected in a new tube. Afterwards, DNA was precipitated using sodium acetate (NaAc, pH 4.8) and isopropanol and dissolved in 35 µL of ultrapure water. The amount and quality of the DNA obtained were evaluated using NanoDrop (ND-1000 Spectrophotometer, NanoDrop Technologies, Wilmington, DE, USA).
Three heterologous microsatellite primers used by Patel et al. (2010) for cross-species amplification of 34 rohu microsatellite loci in Labeo bata and some other cyprinids and a CO1 gene primer designed and used by Ward et al. (2005) for the identification of 207 fish species and assessment of different stocks of farmed L. rohita (Kamran et al., 2023) were selected (Table 1). Primers were synthesized from Macrogen (Macrogen, Seoul, Korea). A 30 µL mixture containing 14 µL of Thermo Scientific PCR master mix (0.05 U/µL Taq DNA polymerase, reaction buffer, 4 mM MgCl2, 0.4 mM of each dNTP), 11 µL PCR water, 1 µL of each primer (forward and reverse) (0.2 µM in a 1 µL volume) (Thermo Fisher Scientific, Waltham, MA, USA) and 3 µL DNA was used in a gradient thermal cycler PCR (BIO-RAD T100TM thermal cycler, Hercules, CA, USA). The following were the conditions used for COI PCR: 1) An initial denaturation at 94°C for 1 minute, 2) Denaturation at 94°C for 60 seconds, 3) Annealing at 54°C for 45 seconds, iv) Elongation at 72°C for 60 seconds, succeeded by 7 minutes at 72°C. There were 35 cycles for denaturation, annealing, and extension steps. Additionally, for amplifying microsatellite primers, the PCR conditions were: i) A 5-minute initial denaturation, ii) Denaturation at 95°C for 1 minute, iii) Annealing of Lr-28 at 58°C, Lr-29 and Lr-37 at 60°C for 45 seconds, iv) Elongation at 72°C for 1 minute and then final elongation at 72°C for 10 minutes. In this PCR procedure, a total of 30 cycles were performed. Finally, the end products were kept at 4°C.
To analyze the amplified products, 1.5% agarose gel (100 mL) solution was prepared by dissolving 1.5 g of agarose powder, 20 mL of 5X TBE buffer (45 mM Tris-borate, 1 mM EDTA), and ethidium bromide was added at a concentration of 0.5 µg/mL visualize the DNA bands. Subsequently, gel electrophoresis was conducted by loading a DNA ladder in one well of each comb, while samples containing 1 µL bromophenol blue dye were loaded in the rest of the wells. After electrophoresis, a few excellent-quality bands were sent to Macrogen (Macrogen, Seoul, Korea) for sequencing.
Manual scoring of the alleles represented as bands on the microsatellite loci was performed, and band sizes were calculated using the DNA fragment program (version 3.03) following the procedure followed by Nash (1991). Determination of various particular-sized alleles at the locus was carried out through genotype scoring for specialized locus.
The sequences underwent alignment via the BioEdit software (Version 7.2.5) (Thompson et al., 1997) and were subsequently identified on the NCBI platform utilizing the BLAST algorithm.
AMOVA was carried out using DnaSP5.exe for the examination of the spatial structure of strains. For both microsatellite and mitochondrial markers, population-wise pairwise FST statistics were computed. The significance levels (p < 0.05) were determined.
To produce a map of the research location, ArcGIS version 10.8 was utilized. GENEPOP version 4.0 (Rousset, 2008) was used to compute several factors, including the frequency and number of alleles/locus (NA), Ho and He, as well as conformity with Hardy-Weinberg Equilibrium (HWE). Allelic richness (Ar) and significance of the FIS within the population and other loci were calculated using FSTAT version 2.9.3.2 (Goudet, 1995) while the difference between and within populations known as FIS was calculated using Genetix version 4.05 software. Moreover, genetic relatedness and variation among strains were determined by constructing a UPGMA dendrogram (Nei, 1972) using the R programming language (version: 3.6.1).
Results
Lr-28, Lr-29, and Lr-37 primers were effectively amplified by PCR, and all three microsatellite loci were identified as polymorphic, with a PIC value exceeding 0.5 (PIC > 0.5). The Lr-28 loci had a PIC value of 0.79, while the Lr-37 loci had a PIC value of 0.59. A total of 12 alleles were recognized at the three loci, with 5 alleles at the Lr-28 loci, 3 alleles at the Lr-29 loci, and 4 alleles at the Lr-37 loci. The number of alleles ranged from 3 to 5, having an average of 4.0 alleles per locus. Overall selected loci (Table 2), allelic size and frequency differed from sample to sample.
The genetic variation observed among different strains is presented in Table 2. The Ho ranged from 0.671 to 0.791 for all strains, with TMs having the highest Ho and TKs having the lowest. Likewise, the He ranged from 0.758 to 0.860, with WKs having the highest He, followed by TMs and TKs having the lowest He. TMs showed the highest genetic variability, while TKs showed the lowest compared to other strains, based on the overall mean value of all loci. The FIS value ranged from 0.103 to 0.325, with the lowest average FIS value for TMs followed by WRs and WKs, while TKs showed the highest average FIS value. Moreover, 12 of the 24 tests showed significant deviations from HWE. Genotype frequencies at all twelve loci across 24 tests showed an overall deviation from HWE (Table 2). CMs, TKs, SPs, and USs showed deviation at two loci each, TMs, MKs, and WKs at one locus, while WRs did not show deviation from HWE at any loci.
The study conducted pairwise comparisons between each strain (Table 3) and found significant differences in their genetic differentiation. The FST value indicated that all strains showed differentiation. The FST value for all strains had an average range of 0.002 to 0.059, indicating a moderate to low level of population structure. The study revealed that the gene flow was highest between WKs and WRs and lowest between CMs and MKs. The highest FST value between CMs and MKs indicated that both strains do not share the allele and are remarkably different (FST = 0.059) while the lowest FST value between TMs and MKs demonstrated maximum gene flow between both strains (FST = 0.002). Similarly, the genetic distance analysis based on the CO1 gene revealed the highest gene flow between WKs and WRs (0.004) while the minimum gene flow between WRs and USs (0.068).
The molecular variance analysis presented in Table 4 revealed a minor genetic differentiation among strains using microsatellites (FST = 0.03524, p = 0.000) and COI gene (FST = 0.04464, p = 0.000). However, the observed genetic variation in the populations (microsatellite marker, 68.98%, and COI gene, 59.36% are at the individual level within the strains).
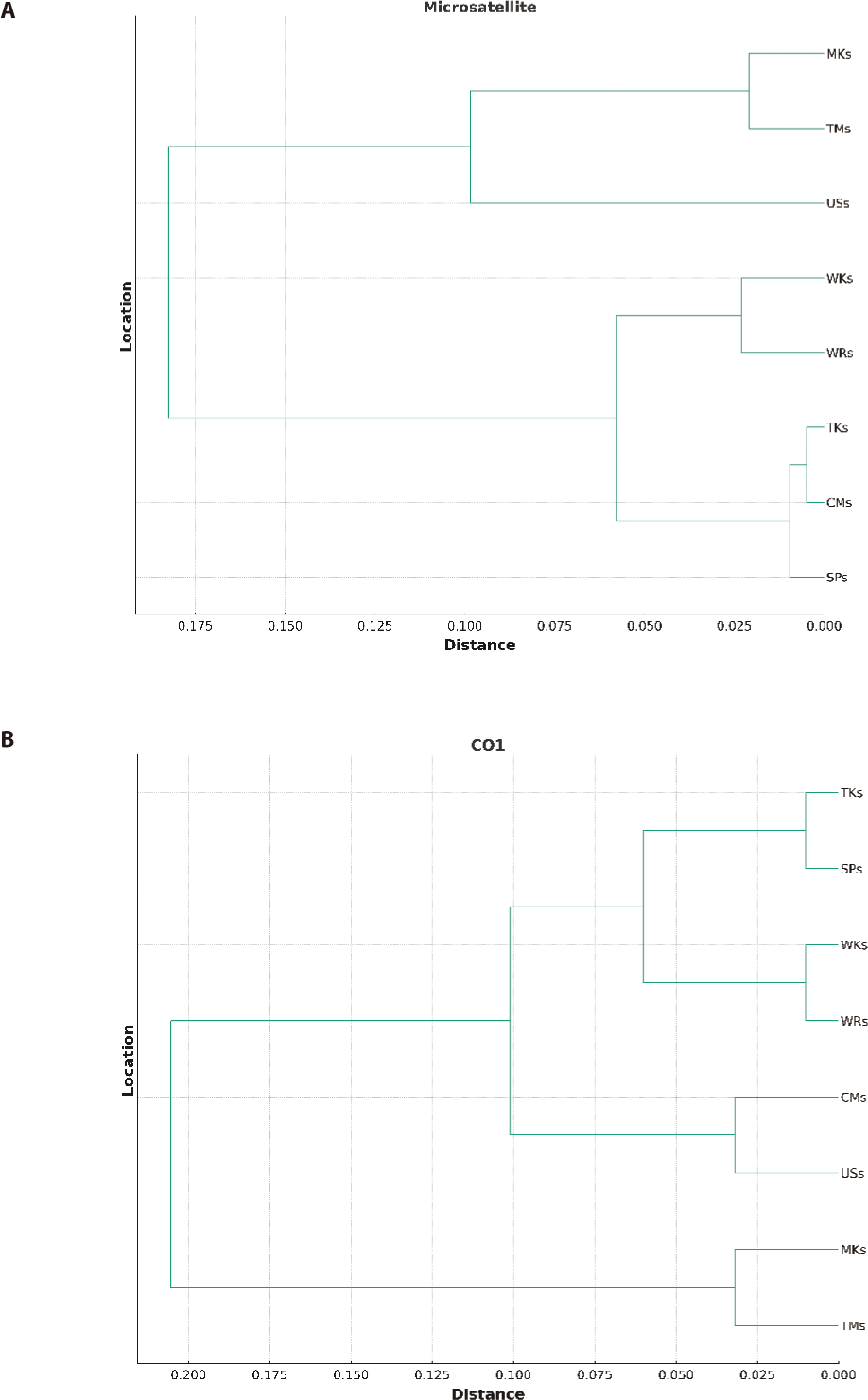
The UPGMA dendrogram, illustrated in (Fig. 2) and based on Nei’s genetic distance (1972), demonstrated the genetic relatedness among all strains. The formation of two major clusters was depicted in Fig. 2A and 2B based on both microsatellite and mtDNA markers. One cluster comprised three strains, while the other consisted of five strains. Sub-clusters were further divided into each cluster. In the first sub-cluster, only the USs strain was present while TMs and MKs were in the other sub-cluster. The second sub-cluster was further divided into two clusters. One cluster had two strains, WRs, and WKs, while the other three strains, CMs, TKs, and SPs, were in the other.
Figures S1 to S4 (supplementary material) show gel electrophoresis images of 680 bp PCR products of mtDNA COI and PCR products of microsatellite markers. The result indicated no modifications (insertion or deletion).
Discussion
To establish efficient conservation methods, improved knowledge of population dynamics, and implementation of a genetic improvement plan for any species and assessment of genetic diversity is a crucial first step. In recent years, microsatellite markers and mtDNA have become frequently used for this aim (Costa-Urrutia et al., 2012). In the current study, three microsatellite markers and an mtDNA marker provided valuable information about the diversity among two wild (WKs & WRs) populations caught from River Kabul and River Ravi respectively, and 6 captive-bred (TMs, TKs, CMs, MKs, SPs, and USs) populations of indigenous culturable species L. rohita collected from different fish hatcheries across Pakistan.
Detecting polymorphisms is crucial for selecting molecular markers. The informativeness and usefulness of a genetic marker in genetic diversity analysis are often measured using the PIC value (Serrote et al., 2020). The selected microsatellite markers in this study were found to be polymorphic, with 12 alleles across three loci and a PIC value of more than 0.5 (0.59–0.79) (Table 2). Genetic evaluation studies frequently prefer microsatellites with 3-4 alleles per locus and a PIC value greater than 5 (Lie et al., 2010). These results suggest that the microsatellite markers used in this study are meaningful for assessing the genetic diversity of L. rohita strains. Previous research has used Lr family markers in combination with other markers to examine population structure and genetic diversity in both wild and farmed L. rohita populations, and similar patterns of genetic variation have been found (Alam et al., 2009; Hussain et al., 2021).
Maintaining genetic variation in a population is the most important in conservation and genetic improvement programs. Genetic diversity allows the population to face environmental challenges in the future and respond to long-term selection, either natural or artificial for desirable traits (Sharma et al., 2016). However, loss of genetic diversity in the captive population due to poor management, lack of technical knowledge, and genetic awareness among fish seed-producing persons is the major issue in Pakistan (Riaz et al., 2023). Though genetic variability of the wild population forms the hub for the selection of stock for the selective breeding program, however, the natural stocks of most of the fish species are under threat due to anthropogenic intervention (Islam & Alam, 2004). Generally, genetic degradation resulted in a decline in fish and consequently affected the sustainability of the aquaculture business. Thus, it is imperative to address genetic problems, genetically characterize the population, and provide baseline data for the initiation of the selective breeding program and supply of quality fish seed.
The most effective approach to measuring genetic variation in populations is the Ho per locus which has a non-random variation not only at the loci level but also at the species and population level (Allendorf & Utter, 1979). The mean value of Ho for all strains ranged from 0.671 to 0.791, with TMs displaying the highest Ho and TKs exhibiting the lowest. Similarly, the mean value of He ranged from 0.758 to 0.860, with WKs demonstrating the highest He, followed by TMs, and with TKs having the lowest value. Here the Ho and He values 0.755–0.766 and 0.829–0.860 respectively of the wild population collected from River Ravi and River Kabul were higher than Ho = 0.655–0.705, He = 0.702–0.725 reported by Nabeela et al. (2014) in the wild population of L. rohita collected from three major rivers (Halda, Jamuna, and Padma) of Bangladesh, indicating higher heterozygosity. Nonetheless, the He (0.5624) recorded in L. rohita specimens collected from three distinct regions of the Chenab River by Hussain et al. (2021) was inferior to the value reported in the current investigation.
In captive populations, small population size and negative selection are the major factors contributing to inbreeding depression and genetic diversity (Sah et al., 2018). However, in the present study, the observed and expected heterozygosity of all populations collected from different fish hatcheries were Ho = 0.67–0.79, He = 0.758–0.85 with the highest in TMs and the lowest in TKs, indicating a somewhat similar level of heterozygosity in some captive population. However, Sultana et al. (2015) reported a low level of Ho = 0.42–0.7 and He = 0.661–0.717 in L. rohita samples collected from six fish hatcheries other than those reported here. In the present study, samples were collected across Pakistan while Sultana et al. (2015) selected hatcheries from only one province. The variation in genetic variability among captive stock may indicate the level of genetic awareness, technicality, and the number of broodstock used for induced breeding. It is a personal observation that TMs had a healthy broodstock and educated/trained staff and are in the practice of rotation of broodstock compared to Tanda Hatchery.
The lower Ho than He in both wild and captive populations in the current study, indicating a departure from HWE and the possibility of inbreeding in successive generations, may be due to non-random mating (Sharma et al., 2016). These deviations were not systematic, detected in 12 out of 24 tests and at different loci for different populations. For instance, WRs did not show deviation at any loci while others deviated at one locus (TMs, MKs, WKs), others at three loci (CMs, SPs, USs), or all loci (TKs). Departure from HWE of a somewhat low magnitude (41.66) has been reported by Alam et al. (2009) in three wild populations while Ullah et al. (2015) detected 61.11% deviation as compared to the observed (50%) in the present study. The difference may be attributed to variable genetic structures (Hussain et al., 2021).
The observed difference between Ho and He was also evident in the positive FIS value in analysis based on microsatellite markers which revealed variation among strains as it ranged from 0.103 (TMs)–0.325 (TKs). Moreover, except TMs all hatchery stocks showed high FIS (0.192–0.325) compared to wild stocks (0.164-0.165). The high positive FIS value is attributed to non-random mating resulting in inbreeding. Like present observations, some other studies also indicate a similar level of FIS value and loss of heterozygosity in the Pakistani stocks of L. rohita (Qadeer & Abbas, 2017; Sultana et al., 2015) and confirming the outcomes of the present study.
The FST analysis between different population pairs based on both microsatellite and mtDNA analysis indicated the low genetic differentiation among different strains and ranged from 0.002 to 0.059 and 0.004 to 0.068 respectively. Both markers indicated the lowest genetic difference and the highest gene flow among the wild population (WKs and WRs). This could be due to the physical connectivity of both rivers as River Kabul meets River Indus at Attock, Punjab, and River Ravi meets River Indus at Mithankot, Rajanpur, Punjab which enables the genetic mixing of the populations despite having a geographical distance in between. Moreover, the highest genetic difference among the hatchery-reared population could be due to the minimum gene flow between these strains because of captive breeding or may be related to geographical isolation (Alam et al., 2009). However, the low genetic distance between MKs and TMs and among CMs, TKs, and SPs could be related to the sharing of brooders between the hatcheries, as Tawakkal Fish Hatchery and Mianchannu Fish Hatchery are in the same Province, Punjab. Similarly, Carp Hatchery and Training Center, Peshawar, Government Fish Hatchery, Charbanda, Mardan, and Tanda Government Fish Hatchery Kohat are situated in the Province of Khyber Pakhtunkhwa. There is a common practice of sharing brooders between hatcheries of the same province instead of another province because of geographical distance. The present investigation affirms the correlation between genetic and geographical separation, which is in line with the previous research conducted by Sultana et al. (2015) and Kamran et al. (2023), who observed a comparable degree of genetic resemblance among populations from three separate hatcheries located in the same province (Faisalabad, Farooqabad, and Mianchannu hatcheries). The UPGMA dendrogram classified the eight strains into two primary clusters based on genetic distance, which were subsequently divided into sub-clusters, indicating the genetic proximity of seven strains. Nevertheless, the USs were genetically different from all other strains.
The AMOVA analysis carried out on microsatellite markers and mtDNA indicated that the molecular diversity among the eight strains was mainly due to individual differences within the population and had less to do with variations between populations. Similar findings were reported by Luhariya et al. (2012) in the wild population of rohu. Both these studies observed that the variations were primarily due to within-individual variations for microsatellite markers and mtDNA gene, respectively. In scattered populations, especially in freshwater species, a higher degree of genetic differentiation is typically expected (Habib, 2010). However, the relatively low genetic diversity observed among the eight L. rohita strains in this study may be attributed to the sharing of ancestral gene pools.
Conclusion
In conclusion, based on both microsatellite and mtDNA markers, all the strains showed intra and inter-strains genetic variation among wild and captive-bred populations of L. rohita, however, the overall variations are low to moderate, reflecting the proper management strategy. Moreover, the results of the current study provide the baseline data for the initiation of a genetic program.
Supplementary Materials
Supplementary materials are only available online from: https://doi.org/10.47853/FAS.2023.e67

