Introduction
Olive flounder (Paralichthys olivaceus) is one of the most farmed fish in South Korea. Its average production over the past five years (2018–2022) was 42,398 tons, accounting for 48.82% of the total production and making it a very important fish species at an industrial level (Statistics Korea, 2024). Although olive flounder aquaculture technology continues to develop, environmental destruction, the emergence of fish pathogens, and instability in fishmeal supply due to the depletion of marine fishery resources negatively impact olive flounder production (Hasan et al., 2018; Lee et al., 2024). In this situation, probiotics are attracting attention as microbial additives that can help build eco-friendly fish farms, improve fish immunity, and enhance the digestibility of low-fishmeal feed (Jang et al., 2023).
Probiotics are defined as live microorganisms that provide beneficial health effects to the host when supplied in adequate amounts (FAO/WHO, 2001). They are considered to aid digestion and absorption, stimulate immune cells, produce antibacterial substances to kill pathogens, and change the composition of the gut microbiota to a healthy one (Jang et al., 2021; Nawaz et al., 2018; Nayak, 2010). However, most probiotics applied to fish so far are lactic acid bacteria of terrestrial origin (Jang et al., 2023; Van Doan et al., 2020). The concept of host-associated probiotics has recently been reported, and attempts have been made to use microorganisms isolated from the intestines of fish as probiotics (Jang et al., 2021, 2022, 2023; Van Doan et al., 2020). However, lactic acid bacteria have also been mainly isolated and targeted for use as probiotics (Cathers et al., 2022; Kader et al., 2021). In fact, lactic acid bacteria, such as Lactobacillus and Lactococcus, are considered beneficial for humans and animals (Jang et al., 2024). However, whether lactic acid bacteria are the optimal probiotic candidates for fish, which have different gut microbiota compositions from humans, remains questionable (Jang et al., 2023; Van Doan et al., 2020). Moreover, the growth temperature of most lactic acid bacteria is similar to the human intestinal temperature. However, because the intestinal temperature of fish is relatively low, it may not be an optimal environment for lactic acid bacteria. Hence, to develop appropriate probiotics, it is important to know the gut microbiota composition of healthy fish.
This study therefore assessed the gut microbiota composition of farmed olive flounder. Gut microbiota analysis was performed by dividing the fish into growth stages according to their weight. Moreover, the microbial composition, relative microbial abundance, and representative microorganisms at each stage were investigated.
Materials and Methods
Ten farmed olive flounders were provided by the Aquafeed Research Center of the National Institute of Fisheries Science (Pohang, Korea). The fish were weighed and divided into a juvenile group weighing less than 100 g and a subadult group weighing between 400 and 800 g (Niu et al., 2020). Then, the intestines of all the fish were aseptically separated. All contents present in the isolated intestines were collected, and the total bacterial DNA in the intestinal contents was isolated using the ZymoBIOMICS DNA Miniprep Kit (Zymo Research, Irvine, CA, USA), according to the manufacturer’s instructions.
The V3–V4 region of the 16S rRNA sequence was amplified from the total DNA of the isolated microorganisms using the forward primer 5′-TCGTC GGCAG CGTCA GATGT GTATA AGAGA CAGCC TACGG GNGGC WGCAG-3′ and the reverse primer 5′-GTCTC GTGGG CTCGG AGATG TGTAT AAGAG ACAGG ACTAC HVGGG TATCT AATCC-3′. PCR was performed with a Veriti 96-well thermal cycler (Applied Biosystems, Foster City, CA, USA) at the Core Facility Center for Tissue Regeneration, Dong-eui University (Busan, Korea). Moreover, 16S rRNA gene amplicon libraries for sequencing were prepared according to the method described in 16S Metagenomic Sequencing Library Preparation: Preparing 16S Ribosomal RNA Gene Amplicons for the Illumina MiSeq System (Illumina, San Diego, CA, USA) (Illumina, 2013).
Paired-end (2 × 300 bp) raw sequencing data were processed using Quantitative Insights Into Microbial Ecology 2 (QIIME2) v2023.5 (https://qiime2.org/) (Bolyen et al., 2019). The 5′ end of the primer was trimmed, and the 3′ end was truncated to the region where the average quality score per sequence location was greater than 25. Sequences were filtered, denoised, and merged and chimeras were removed using the Divisive Amplicon Denoising Algorithm 2 (DADA2) pipeline (GitHub; https://github.com/benjjneb/dada2/); reads were clustered into amplicon sequence variants (ASVs) (Callahan et al., 2016). The ASVs were taxonomically assigned by the sklearn classifier method using SILVA database v138 (arb-silva, Bremen, Germany; https://www.arb-silva.de/). Reads not assigned to a taxon were removed from the analysis by using the taxa filter-table function. The results of alpha diversity analysis, beta diversity analysis, microbial composition analysis, and linear discriminant analysis (LDA) effect size (LEfSe) (Segata et al., 2011) were visualized using QIIME2 View (https://view.qiime2.org/) and MicrobiomeAnalyst (Microbiomeanalyst, Montreal, QC, Canada; https://www.microbiomeanalyst.ca/) (Chong et al., 2020).
Results
On measuring the weight of the olive flounders, five fish were found to weigh less than 100 g; these fish were included in the juvenile group. The average weight of the fish in the juvenile group was 90.30 ± 4.47 g. The remaining five fish were included in the subadult group; their average weight was 505.80 ± 58.48 g.
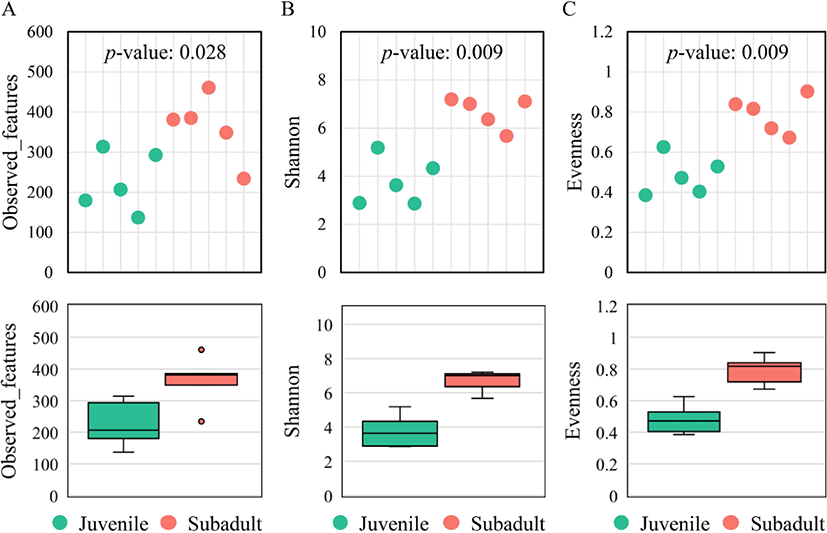
Alpha diversity was compared between the groups based on three estimators: observed features, Shannon’s evenness index, and Pielou’s evenness index. The observed features metric was significantly higher (p = 0.028) in the subadult group (362 ± 82) than in the juvenile group (226 ± 75) (Fig. 1A). Shannon’s and Pielou’s evenness indices were also significantly higher (p = 0.009) in the subadult group (6.67 ± 0.64 and 0.79 ± 0.09, respectively) than in the juvenile group (3.78 ± 0.99 and 0.48 ± 0.10, respectively) (Fig. 1B and 1C).
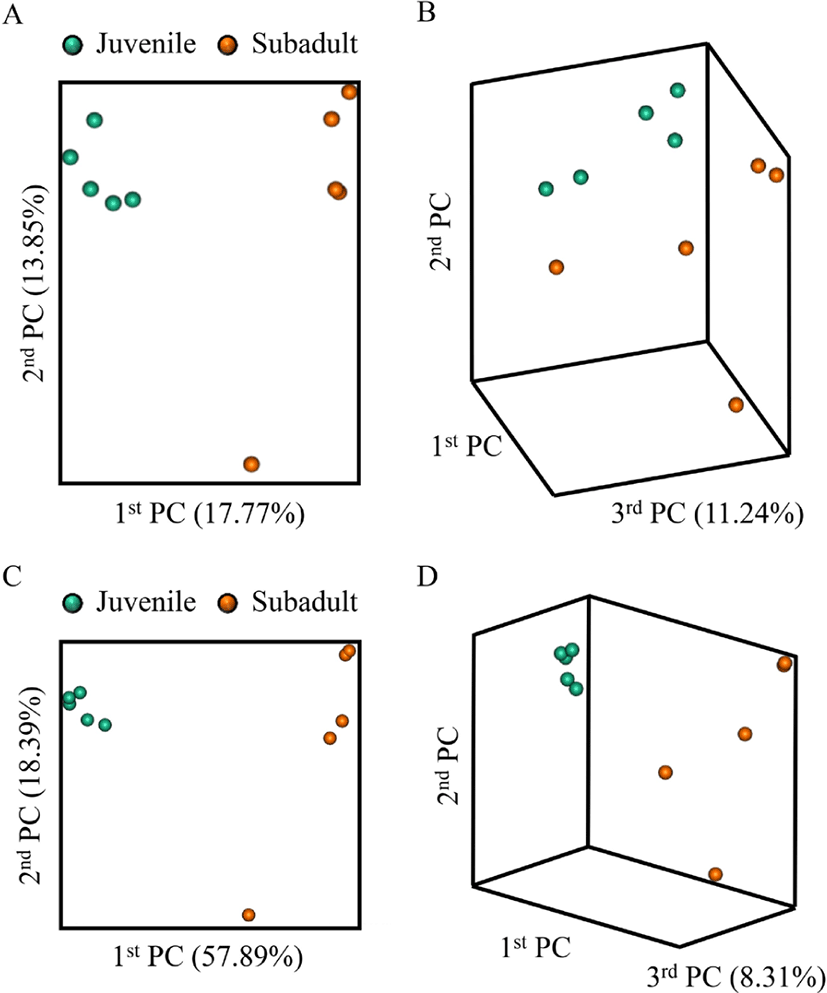
Beta diversity analysis of the olive flounder gut microbiota was performed based on the Jaccard distance (Fig. 2A and 2B) and Bray-Curtis distance (Fig. 2C and 2D) between the samples using the principal coordinate analysis (PCoA) plot. In all the results, the two groups showed clear boundaries, indicating that differences existed between the groups.
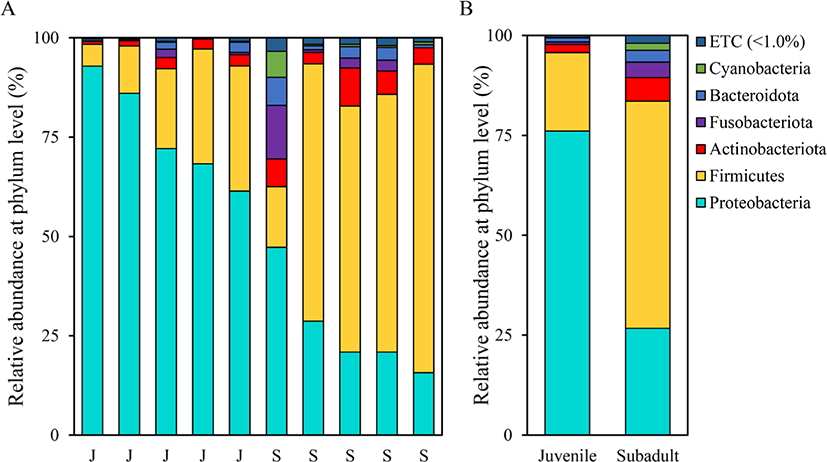
Assessment of the relative microbial abundance at the phylum level based on the SILVA database revealed differences in microbial compositions between the two groups (Fig. 3). The most abundant phylum in the juvenile group was Proteobacteria (76.13 ± 12.98%), followed by Firmicutes (19.58 ± 11.05%) and Actinobacteriota (2.03 ± 0.95%). On the other hand, the most abundant phylum in the subadult group was Firmicutes (56.88 ± 24.06%), followed by Proteobacteria (26.68 ± 12.40%) and Actinobacteriota (5.90 ± 5.51%).
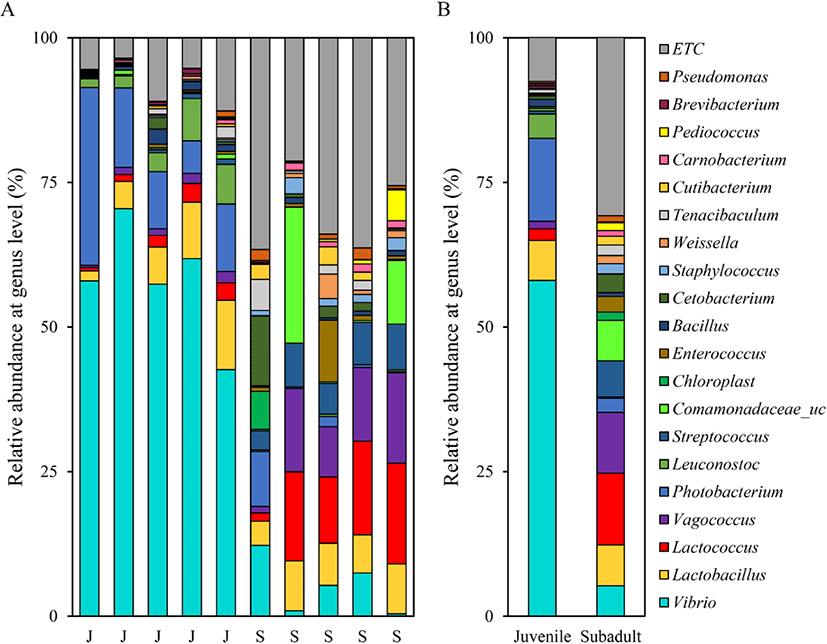
Assessment of the relative abundance at the genus level revealed that the most abundant genus in the juvenile group was Vibrio (58.06 ± 10.09%), followed by Photobacterium (14.33 ± 9.67%) and Lactobacillus (6.93 ± 4.06%). The most abundant genus in the subadult group was Lactococcus (12.37 ± 6.50%), followed by Vagococcus (10.53 ± 5.88%) and Lactobacillus (7.07 ± 1.84%) (Fig. 4).
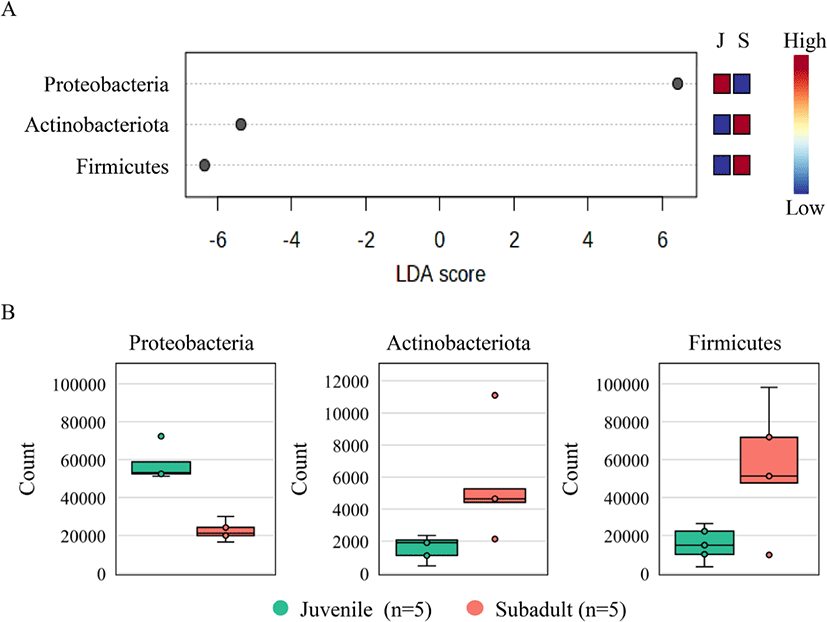
LEfSe revealed a total of three phyla in the juvenile and subadult groups based on an LDA score of 4. The phylum Proteobacteria represented the juvenile group, while the phyla Firmicutes and Actinobacteriota represented the subadult group (Fig. 5A). A comparison of the observed phylum counts in the juvenile and subadult groups revealed that the Proteobacteria counts were 57,571 and 22,337, respectively; Firmicutes counts were 15,352 and 55,717, respectively; and Actinobacteriota counts were 1,580 and 5,516, respectively (Fig. 5B).
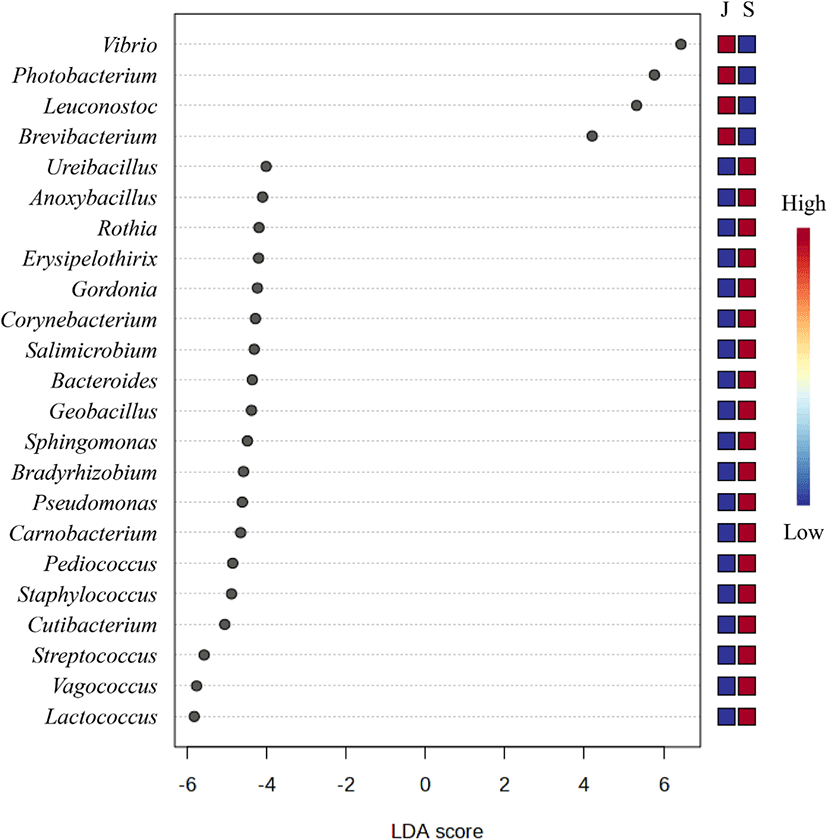
Based on an LDA score of 4, the genera Vibrio, Photobacterium, Leuconostoc, and Brevibacterium were considered to represent the juvenile group. On the other hand, 19 genera, including Lactococcus, Vagococcus, and Streptococcus, represented the subadult group (Fig. 6).
Discussion
A striking feature of vertebrate evolution is that vertebrates have developed adaptive immunity, expressing antigen-specific receptors that allow them to recognize and remember microorganisms (McFall-Ngai, 2007). Studies have suggested that this evolution of vertebrates has promoted the expansion of their resident microbiota (Maynard et al., 2012; McFall-Ngai, 2007). Therefore, while the gut microbiota of invertebrates consists of only a small number of species, that of vertebrates is more complex. Moreover, complex interactions exist between the host and the microbiota (Kostic et al., 2013; McFall-Ngai, 2007). In humans, infants are born with an essentially sterile intestine. Thereafter, dramatic changes can occur in microbial diversity during infancy. However, Bacteroidetes and Firmicutes gradually become prevalent in adults, a composition that is conserved across humans (Kostic et al., 2013). Similarly, in this study, differences were noted in the gut microbiota diversity and composition of olive flounder depending on the growth stage. Alpha diversity analysis revealed that the gut microbiota of the subadult group was more diverse than that of the juvenile group. In addition, the gut microbiota composition at the phylum and genus levels was more diverse and complex in the subadult group.
Phylum-level analysis of the gut microbiota according to the growth stages of olive flounder revealed a clear difference between the groups; the abundance of Proteobacteria decreased, while that of Firmicutes increased. Proteobacteria was the predominant phylum in all fish. Studies using zebrafish have revealed that Proteobacteria can cause intestinal inflammation in fish (Brugman et al., 2009). On the other hand, Firmicutes may have a direct effect on fatty acid storage (Ley et al., 2006, 2008; Turnbaugh et al., 2009). These results indicate that juvenile olive flounder may be prone to intestinal inflammation and that the gut microbiota composition may change to facilitate fat storage as the olive flounder progresses to subadult stages. Interestingly, in our previous study, the most abundant phylum in the gut microbiota of olive flounder, belonging to the juvenile group, was also Proteobacteria. Additionally, supplementation with appropriate probiotics decreased the proportion of Proteobacteria and increased Firmicutes. The group fed probiotics had increased digestive enzyme activity (Lee et al., 2024). However, this interpretation cannot be confirmed until the effects of specific microorganisms are investigated in actual germ-free olive flounder.
In this study, Vibrio and Photobacterium were representative genera in the gut microbiota of juvenile olive flounder. Differences in the gut microbiota composition may exist as a characteristic of the diversity in fish (Egerton et al., 2018). Nevertheless, studies conducted to date have shown that the genera Vibrio, Photobacterium, and Clostridium are key members of the fish gut microbiota (Egerton et al., 2018). Clostridium is associated with herbivorous diets, while Vibrio and Photobacterium are commonly found in carnivores (Egerton et al., 2018). This study confirmed that Vibrio and Photobacterium are abundant in olive flounder, a carnivorous fish species, during the juvenile stage. However, changes are induced as it progresses to subadult stages.
Many studies are attempting to use probiotics to manipulate the gut microbiota composition toward a healthy one (Jang et al., 2019, 2020). However, one of the biggest obstacles is the lack of data on the baseline gut microbiota composition of healthy fish (Monroig et al., 2013). Therefore, increasing the focus on collecting such data is essential to ensure that diet manipulation strategies achieve their full benefit (Monroig et al., 2013). In this study, Vibrio was abundant in the intestines of healthy juvenile flounder. Vibrio species are well known to cause vibriosis (Sanches-Fernandes et al., 2022). However, some of the reported Vibrio species are the main bacterial species producing polyunsaturated fatty acids (PUFAs) in the fish gut (Monroig et al., 2013). Therefore, studying the potential utilization of Vibrio species as PUFA-producing bacteria for probiotic purposes could be a novel and interesting research avenue for fish health and nutrition (Monroig et al., 2013). Similarly, based on the results of this study, the use of microorganisms presents in subadult olive flounder, such as Lactococcus and Vagococcus species, could also be considered.







