Introduction
Gonadotropin-releasing hormone (GnRH) is a neuropeptide hormone secreted in the brain that is involved in a variety of functions in vertebrates and invertebrates during reproduction. In fish reproduction, GnRH acts on pituitary gonadotropes to induce the synthesis and release of gonadotropins such as follicle stimulating hormone and luteinizing hormone into the bloodstream which regulate all gonadal activities, including the onset of gonadal development, germ cell development (oogenesis and spermatogenesis), sex steroid production, ovulation, and spawning (Muñoz-Cueto et al., 2020; Qin et al., 2022; Zohar et al., 2010). Additionally, GnRH also stimulates the release of other hormones such as growth hormone, prolactin, and somatostatin (Qin et al., 2022; Weber et al., 1997). In vertebrates, GnRH is found in three forms; GnRH1, GnRH2, and GnRH3, where all vertebrates have at least two forms of GnRH. In fish, there are two or three forms of GnRH that consists of differences not only in the order of their amino acid sequences, but they are also differences in embryonic origins, localizations, and functions (Amano et al., 1997; Dubois et al., 2001; Muñoz-Cueto et al., 2020; Selvaraj et al., 2021). Previous studies stated that GnRH2 and GnRH3 were discovered early on the first day after fertilization and GnRH1 expression appeared significantly later (González-Martinez et al., 2002; Zohar et al., 2022).
This study focuses on two potential pangasius candidates: striped catfish, Pangasianodon hypophthalmus (Sauvage 1878), this species is presently cultivated and distributed to over 100 countries globally, and silver catfish, Pangasius nasutus (Bleeker 1863), a native freshwater fish species that is only found in Malaysia and Southeast Asia, where this species is typically captured in the wild and not cultured yet due to the lower survival rate in captivity (Tarkan et al., 2020; Zulkiflee et al., 2020). High demand for this species has encouraged the application of commercial hormone in induced breeding of these species to increase the production to meet the market demand. However, some commercial fish especially wild fish failed to induce breeding in captive environment. This inability to reproduce in captivity due to several environmental and social differences between the wild and captive environments have disrupted hormonal control at the level of the hypothalamus-pituitary-gonadal axis as opposed to natural endocrine variations in the wild (Hassin et al., 1997; Milla et al., 2021).
The fact that many economically significant finfish do not spawn spontaneously in captivity led to the development of efficient and cost-effective artificial hormone-based GnRH for induced breeding. Previous findings report that the use of salmon gonadotropin-releasing hormone (sGnRHa) with a combination of dopamine antagonist (DA) successfully stimulated ovulation in cyprinids and catfish species (Rahdari et al., 2014). Other studies reported that a single injection of 20 μg/kg GnRHa could induce ovulation in tench (Tinca tinca), whereas two injections of 20 μg/kg GnRHa combined with DA were required to induce spawning in grey mullet (Mugil cephalus) (Aizen et al., 2005; Mehdi & Mousavi, 2011). Although the application of commercial hormones offers benefits in terms of potency and guaranteed breeding responses, it is still limited in terms of efficiency in some species which resulting in a varying success rate and the possibility of spawning failure.
Information on the reproductive hormones of P. nasutus and P. hypophthalmus is still limited, as there has been little research on the reproductive biology of these species and none has focused on the molecular characterization of the GnRH genes. Therefore, this study aimed to determine the molecular characterization and phylogenetic classification of GnRH subunit genes in P. hypophthalmus and P. nasutus. The findings of this study could initiate the development of a hormone therapy that is potentially more effective in aiding reproductive failure in P. hypophthalmus, P. nasutus, and other catfish species in captivity. The DNA sequences could be utilised to develop recombinant artificial hormones specifically for catfish species with a suitable biological function in stimulating gonad development, final oocyte maturation, and spermiation.
Materials and Methods
This research was conducted in an ethical and accordance with the code of ethics, following guidelines for the care and use of animals. All procedures were performed following the ethical rules and protocol approved by the Institutional Animal Care and Use Committee of Universiti Putra Malaysia (IACUC Reference Number: UPM/IACUC/AUP-U027/2022).
Three prepubertal of P. nasutus (± 400–500 g) were obtained from Three Ocean Fishpond & Trading Sdn Bhd Rawang, Malaysia and three prepubertal of P. hypophthalmus (± 500–600 g) were obtained from Aquaculture Research Centre Puchong, Universiti Putra Malaysia. The fish were euthanized using MS-222 (Sigma-Aldrich, St. Louis, MO, USA). Brains samples were collected from each fish, immersed in RNAlaterTM solution (Ambion, Austin, TX, USA) and immediately stored at –80℃ until further analysis.
Total RNA was isolated from each tissue using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. The extracted RNA was resuspended in nuclease-free water. Total RNA concentration was determined using Nanodrop spectrophotometer (Thermo Scientific, Madison WI, USA) to measure absorbance at A260nm/A280nm. Next, by using QuantiTect® Reverse Transcription Kit (Qiagen, Hilden, Germany), 1 μg of total RNA was reverse-transcribed (RT) into complementary DNA (cDNA) to serve as a template for polymerase chain reaction (PCR) amplification. The first strand of cDNA synthesis was carried out in a volume of 20 μL reaction mixture (5X Quantiscript RT buffer, Quantiscript Reverse Transcriptase, RT Primer Mix, and RNA) and incubated using Mastercycler Gradient (Eppendorf, Hamburg, Germany) according to the manufacturer’s instructions.
Primers for full-length sequences of GnRH1 and GnRH2 were designed using OligoAnalyzerTM Tool available via online (https://sg.idtdna.com/calc/analyzer) based on the predicted P. hypophthalmus GnRH1 and GnRH2 genes (Genbank accession numbers XM_026924976.1 for GnRH1 and XM_026929077.2 for GnRH2). For GnRH3, primers were designed based on conserved regions of catfish species, due to the unavailability of GnRH3 sequence of the predicted P. hypophthalmus in GenBank. For an effective DNA amplification, the selected forward and reverse primers were analysed to avoid any potential hairpin and primer-dimer interactions formed during PCR amplification. List of primers used in this study is presented in Table 1.
Polymerase chain reaction (PCR) was performed to amplify the cDNA encoding GnRH1, GnRH2 and GnRH3 of P. hypophthalmus and P. nasutus using previously designed primers. The PCR mixture was prepared in 25 μL final volume using PCR master mix (Promega, Madison, WI, USA) containing nuclease-free water, 5X green GoTaq® reaction buffer, GoTaq® DNA polymerase, 10 mM of each dNTPs, 25 mM MgCl2, 1 ng DNA templates, and 20 μM forward and reverse primers. PCR amplification started with initial denaturation for 3 minutes at 95℃, followed by 35 cycles of denaturation at 95℃ for 1 minute, annealing at 54℃–59℃ and extension at 72℃ for 1:30 minutes. The elongation phase was prolonged to 10 minutes at 72℃. The amplified PCR products were separated on a 1.5% agarose gel, stained with DiamondTM Nucleic Acid Dye (Promega), and visualised under UV light. The selected fragments of PCR product were purified using a GeneJET Gel Extraction Kit (Thermo Scientific).
The purified PCR product was cloned into a vector using TOPO® TA Cloning kit (Invitrogen) following protocols given by the manufacturer. The ligated product was transformed into competent cell E. coli, TOP 10 (Invitrogen) following heat-shock process at 42℃ for 30 second and was quickly transferred into ice to complete the transformation process. Later, 250 µL of a pre-warmed super Optimal Culture medium was added, and the tube was shaken horizontally at 200 rpm for an hour at 37℃. Finally, approximately 10–50 µL of the transformed cells was grown overnight at 37℃ on Luria-Bertani (LB) agar (Merck, Darmstadt, Germany) added with 50 μg/mL ampicillin (Sigma-Aldrich) and 40 μg/mL of X-gal (MilliporeSigma, Burlington, MA, USA).
White colonies of positive recombinant clones were chosen for PCR screening using specific primers. The positive colonies were grown in LB broth (Merck) added with 50 μg/mL ampicillin and incubated at 37℃ with agitation at 200 rpm. The bacteria culture was extracted by centrifuging it at 12,200×g for 5 minutes, followed by plasmid extraction using the GeneJET Plasmid Miniprep kit (Thermo Scientific) following manufacturer’s protocols. The DNA samples were then sent to FirstBase Laboratories in Malaysia for plasmid sequencing using universal primers, M13F (–20) and M13R (–20).
The nucleotide sequences were analyzed using the BLASTn, Basic Local Alignment Search Tool, provided by the NCBI (National Center for Biotechnology Information, Bethesda, MD, USA) website at https://blast.ncbi.nlm.nih.gov/Blast.cgi. Multiple alignment was performed to compare similarities between GnRH1 and GnRH2 of P. hypophthalmus and P. nasutus, with other fishes and vertebrates. The similarities of amino acids were also evaluated by implanting ClustalW and calculating the distance matrix. The phylogenetic trees of the two genes, GnRH1 and GnRH2, were constructed using the neighbor-joining method with MEGA11 (Molecular Evolutionary Genetics Analysis) software. The phylogenetic tree of protein was constructed, and bootstrap value was calculated over 1,000 replicates to evaluate the robustness of the inferred nodes.
Results
In the present study, full-length cDNA of GnRH1 and GnRH2 genes of striped catfish, P. hypophthalmus, and silver catfish, P. nasutus were successfully cloned into TOPO TA vector. Based on Fig. 1, the full-length of GnRH1 and GnRH2 were detected in both species at 300 bp and 360 bp, respectively. However, no band was detected for GnRH3 gene in both species.
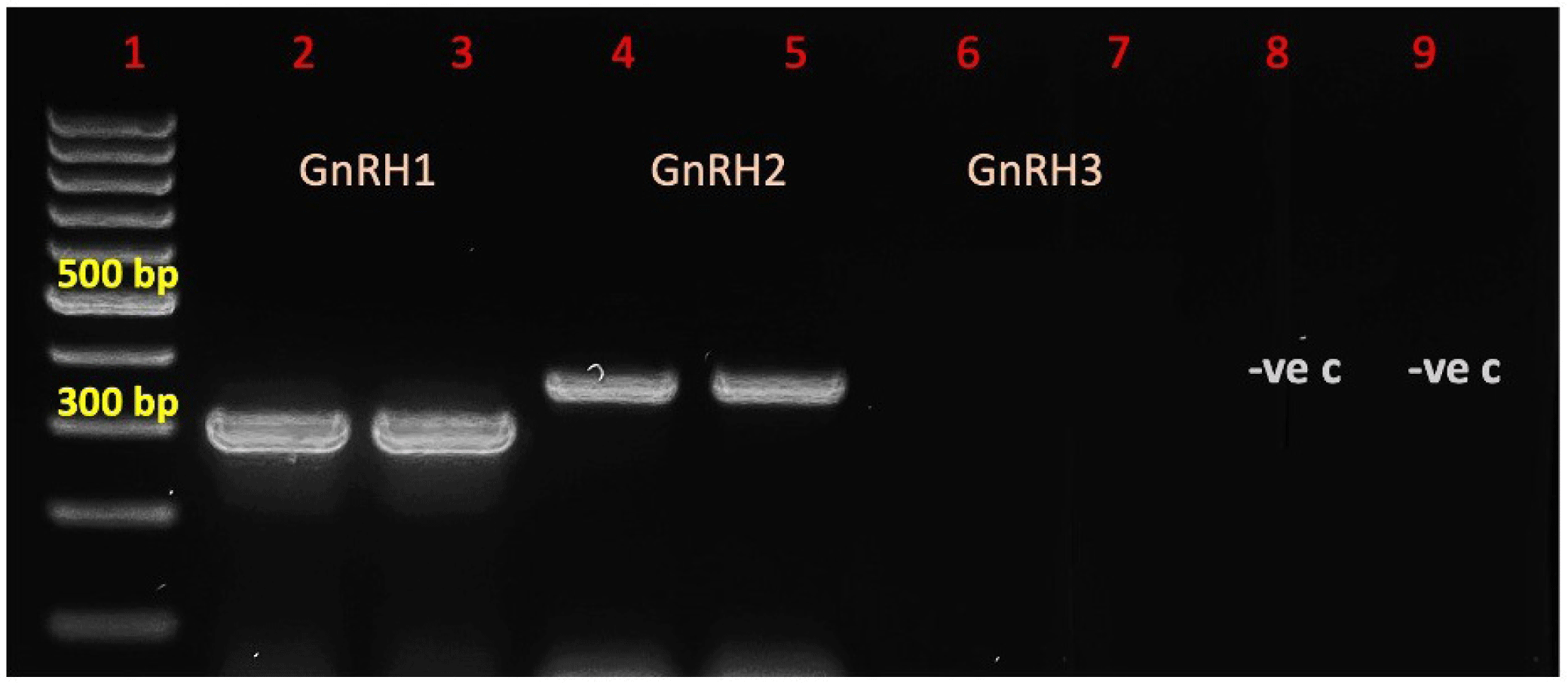
For the GnRH1 gene of P. hypophthalmus, BLASTn result showed P. hypophthalmus shared the highest percentage identity 95.38% similar with P. nasutus, followed by 91.45% similar with Ictalurus furcatus, 90.26% similar with S. meridionalis and 88.34% similar with Hemibagrus nemurus. Meanwhile, for the GnRH2 gene of P. hypophthalmus, BLASTn result showed P. hypophthalmus has the highest percentage identity 93.14% similar to P. nasutus, followed by 91.50% similar with Tachysurus fulvidraco, 91.19% similar with Ictalurus punctatus and 88.89% similar with Clarias gariepinus.
For the GnRH1 gene of P. nasutus, BLASTn result showed P. nasutus has the highest percentage identity 96.26% with P. hypophthalmus, followed by 92.81% similar to I. punctatus, 88.12% similar with S. meridionalis, 88.52% similar with H. nemurus and 84.77% similar with C. gariepinus. Meanwhile, for GnRH2 gene of P. nasutus, BLASTn result showed P. nasutsu has the highest percentage identity 97.84% with P. hypophthalmus, followed by 91.69% similar with I. punctatus, 91.03% similar with H. nemurus, 90.28% similar with T. fulvidraco and 90.20% similar with C. gariepinus.
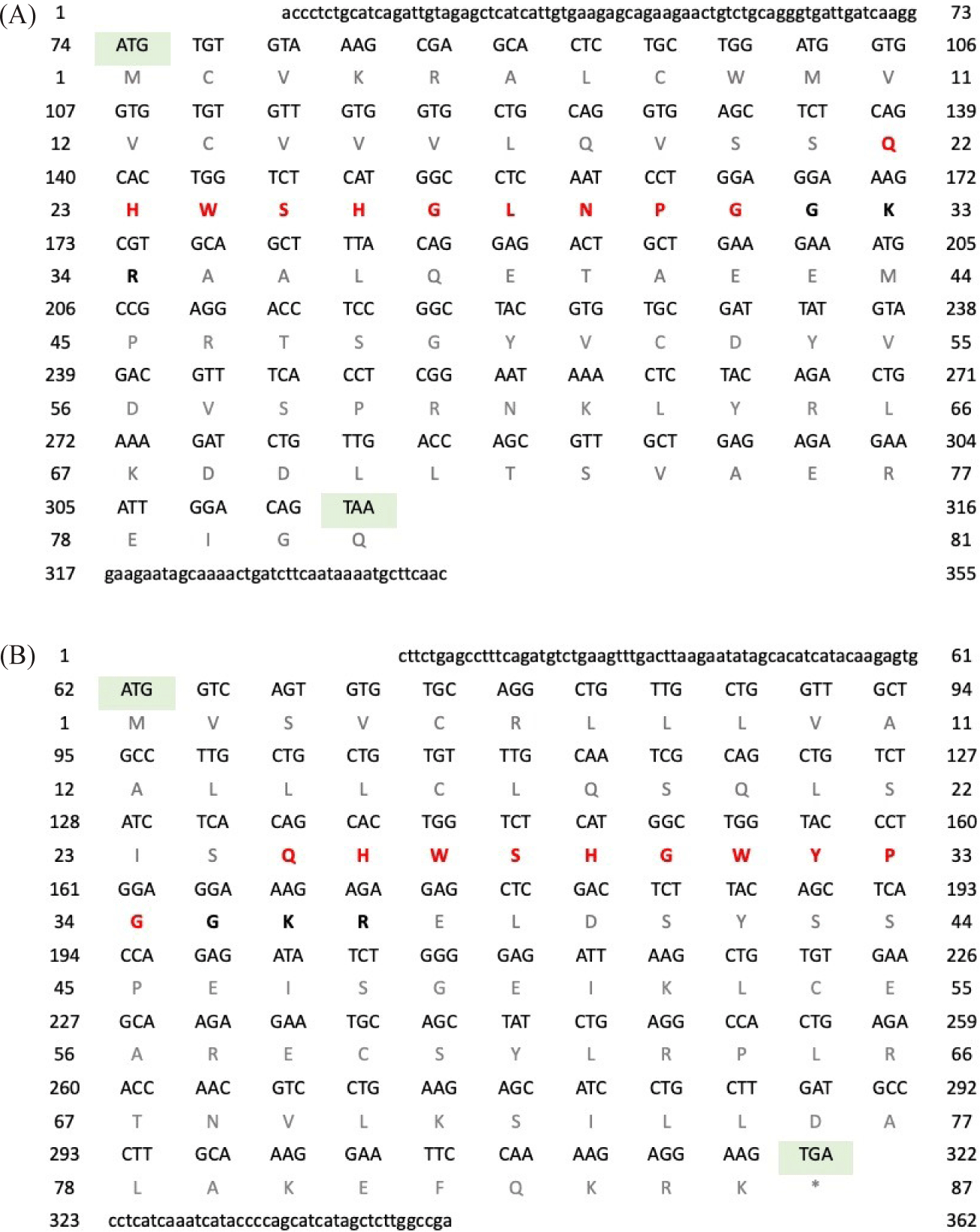
For P. hypophthalmus, the full-length cDNA of GnRH1 was 357 bp (Fig. 2A); consist of 76 bp of 5′ untranslated region (UTR), 38 bp of 3′ UTR with a polyadenylation signal (AATAAA), and 243 bp of an open reading frame (ORF), encoding a protein precursor of 81 amino acids (aa). Amino acids sequence analysis revealed that the prepro-GnRH1 comprised 21 aa of predicted signal peptide, 10 aa of GnRH1 decapeptide, 3 aa of a proteolytic cleavage site, and 47 aa of GnRH1 associated peptide (GAP). Meanwhile, the full-length cDNA of GnRH2 was 382 bp long (Fig. 2B); consists of 64 bp 5′ UTR, 57 bp 3′ UTR, and 261 bp of an ORF, encoding precursor of 87 amino acids. Amino acids sequence analysis revealed that the prepro-GnRH2 comprised 24 aa of predicted signal peptide, 10 aa GnRH1 decapeptide, 3 aa of a proteolytic cleavage site, and 50 aa of GAP. Both GnRH1 and GnRH2 genes of P. hypophthalmus were deposited into GenBank with accession no. OQ685894 (GnRH1) and OQ685895 (GnRH2).
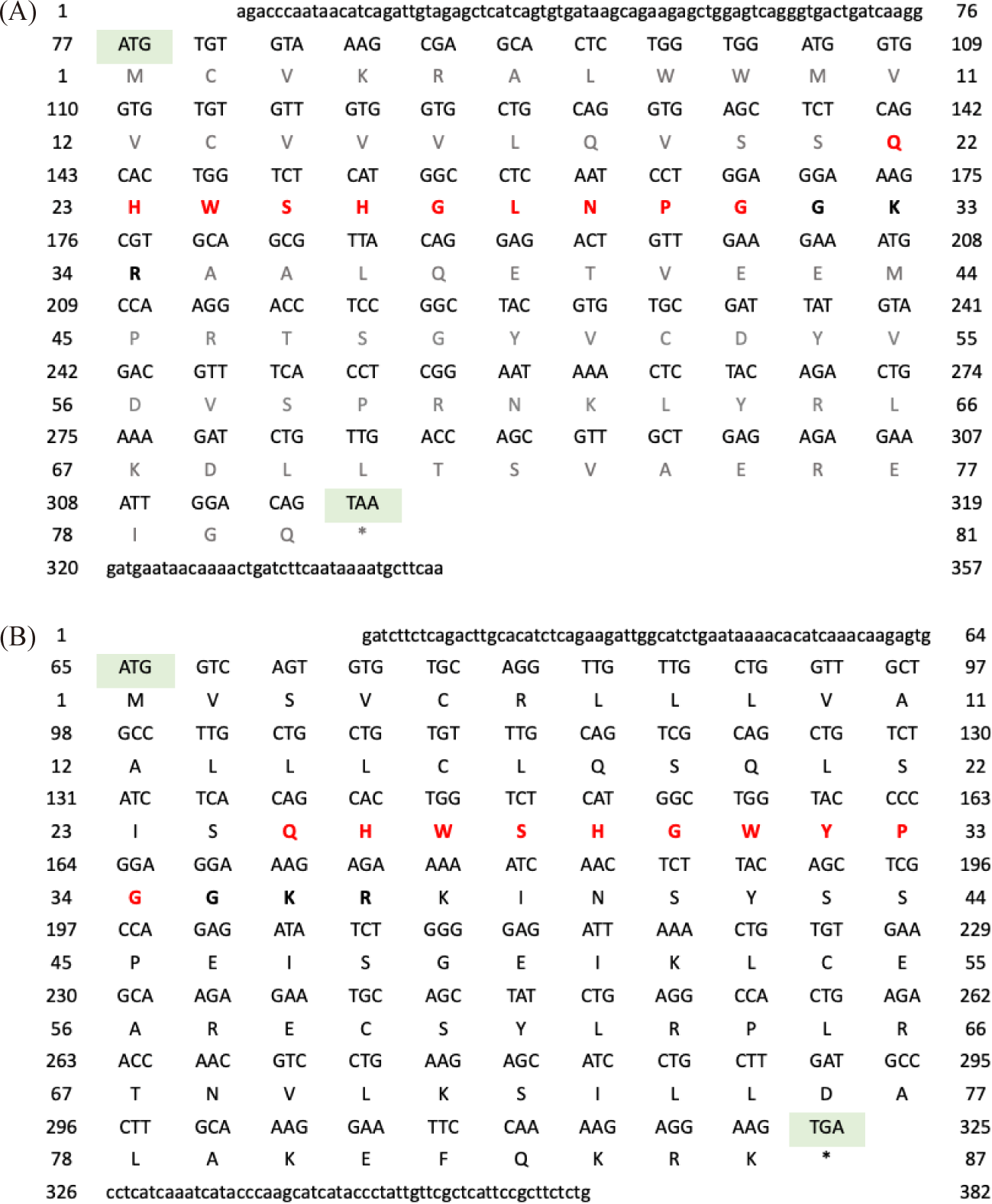
For P. nasutus, the full-length cDNA of GnRH1 was 355 bp long (Fig. 3A); consists of 73 bp of 5’ UTR, 39 bp of 3’ UTR with the polyadenylation signal sequence (AATAAA) and 243 bp of an ORF, encoding precursor of 81 amino acids. Amino acids sequence analysis revealed that the prepro-GnRH1 comprised of 21 aa of predicted signal peptide, 10 aa GnRH1 decapeptide, 3 aa of a proteolytic cleavage site, and 47 aa of GAP. Meanwhile, the full-length cDNA of GnRH2 was 362 bp long (Fig. 3B); consists of 61 bp 5′ UTR, 40 bp 3′ UTR, and 261 bp of an ORF, encoding precursor of 87 amino acids. Amino acids sequence analysis revealed that the prepro-GnRH2 comprised 24 aa of predicted signal peptide, 10 aa GnRH1 decapeptide, 3 aa of a proteolytic cleavage site, and 50 aa of GAP. Both GnRH1 and GnRH2 genes of P. nasutus were deposited into GenBank with accession no. OP104320 (GnRH1) and OP104321 (GnRH2).
Multiple alignments showed that prepro-GnRH in catfish species has a signal peptide, the GnRH decapeptide, the cleavage site, and the GnRH associated peptide region (Figs. 4A and 4B). Comparing the signal peptide and GAP region of GnRH1 revealed a highly conserved sequence region between catfish species, but highly variable sequences between different orders of fish and vertebrates, including perciformes, cyprinodontiformes, clupeiformes, anguilliformes, anura, rodentia, and primates. Multiple sequence alignments showed that the GnRH1 decapeptides structure pGlu-His-Trp-Ser-His-Gly-Leu-Asn-Pro-Gly-NH2 (QHWSHGLNPG) are highly conserved in Pangasius sp. and other siluriformes species. The GnRH1 decapeptide sequence is fairly conserved at positions 1–4, 6, 7, 9, and 10, with one or two variations at positions 5 change to Tyr or Phe and at position 8 changed to Ser or Arg in GnRH1 decapeptides of other vertebrates (Fig. 4A).
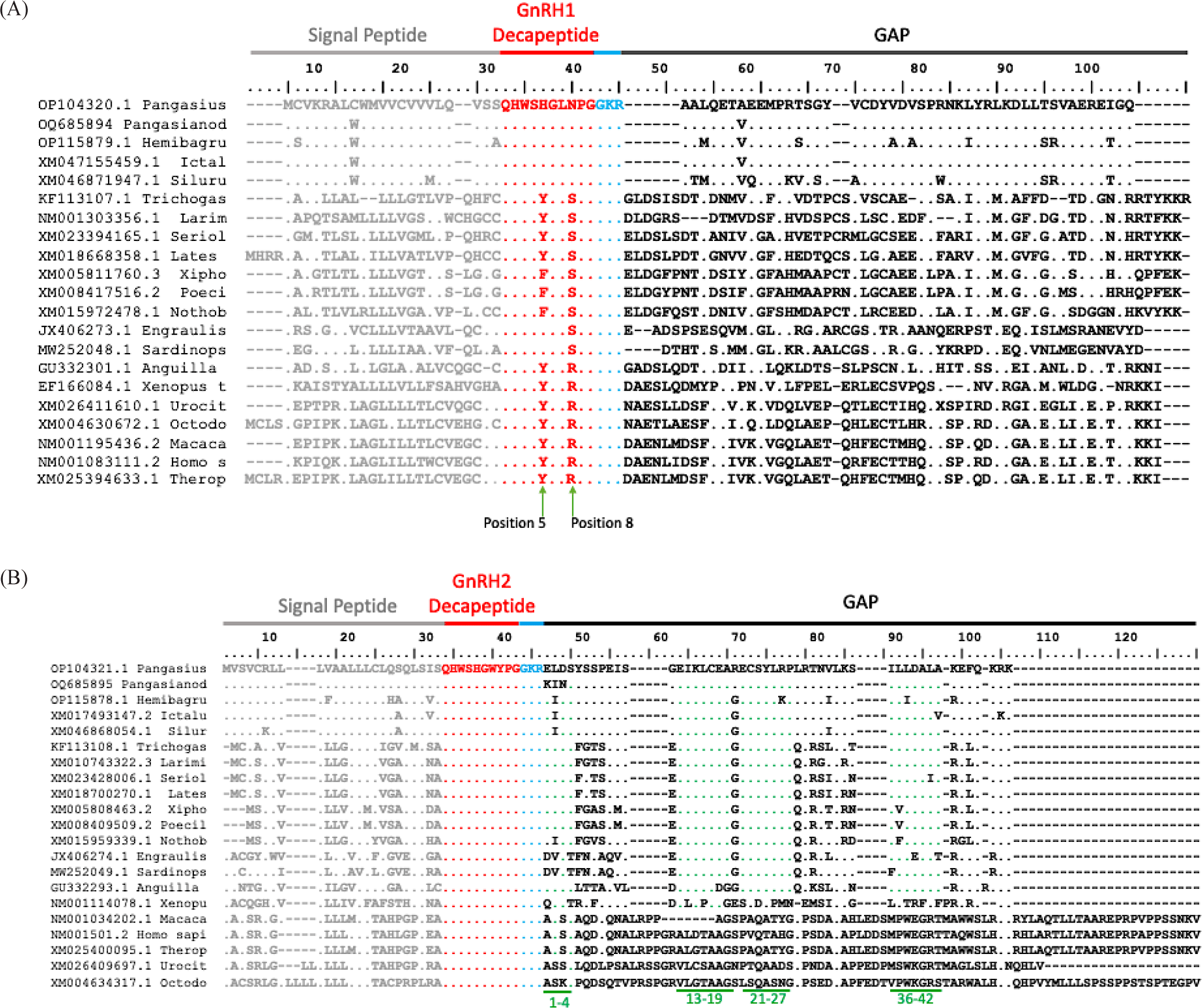
Meanwhile, the GnRH2 decapeptide structure pGlu-His-Trp-Ser-His-Gly-Trp-Tyr-Pro-Gly-NH2 (QHWSHGWYPG) is highly conserved throughout vertebrates for the prepro-GnRH2 gene without any modification (Fig. 4B). The similar result was obtained when comparing the signal peptide and GAP region of catfish species with other vertebrates, as there is a highly conserved sequence of signal peptide within siluriformes order and substantially varied sequences when compared to other vertebrates. However, in the GAP region (position 1–4, 13–19, 21–27, 36–42), GnRH2 sequences demonstrated a well-conserved region not just in the siluriformes order but also within other fishes order, but the GAP region was varied when compared to amphibia and mammalia.
Sequence comparison of GnRH peptides among vertebrates showed that P. hypophthalmus and P. nasutus for GnRH1 and GnRH2 shared high amino acid similarity with their homologs (Table 2). P. hypophthalmus and P. nasutus prepro-GnRH shared high level of sequence identitiy for GnRH1 and GnRh2 with its homology (> 99.7%–100% identity) to the GnRH decapeptide of H. nemurus (river catfish), I. punctatus (channel catfish), and S. meridionalis (southern catfish). A similar result was observed for GnRH2, with catfish from the same order sharing the highest percentage of identities (> 99.6%–99.9% identical) when compared to other vertebrates. When comparing GnRH1 and GnRH2 genes from different classes and orders, from fish to mammalian, the degree of sequence similarity appears to be decreasing in percentage identity. When comparing GnRH1 and GnRH2 across vertebrates, the results showed a similar decreasing sequence identity trend. The lower number of sequence identities can be explained by the less conserved regions of peptide structure between its order and peptide sequence, which are greatly variable.
Phylogenetic tree was built to illustrate the evolutionary relationship between the GnRH genes in P. hypophthalmus, P. nasutus and other vertebrates. The phylogenetic tree of GnRH1 (Fig. 5A) segregated into two separated clades and further separated into two sister groups. According to multiple alignments and the phylogenetic tree, we identified the tree was formed according to the differences in the GnRH1 decapeptide. The first clade formed into two sister groups, with the first sister group having the GnRH1 decapeptide sequence QHWSYGLSPG/QHWSFGLSPG with the highest bootstrap value (100%) and consisting of perciformes and cyprinodontiformes, while the second sister group has the decapeptide sequence QHWSYGLRPG and consists of the orders anguilliformes, anura, rodentia, primates, and clupeiformes. The second clade consist of P. hypophthalmus and P. nasutus, aligned closely with other catfish species, including the H. nemurus (river catfish), I. punctatus (channel catfish), and S. meridionalis (southern catfish), which is consistent with the result of amino acid sequence comparison. Clupeiformes cluster together in one root with siluriformes due to the fairly conserved GnRH1 decapeptide, as can be seen in multiple alignment (QHWSHGLRPG).
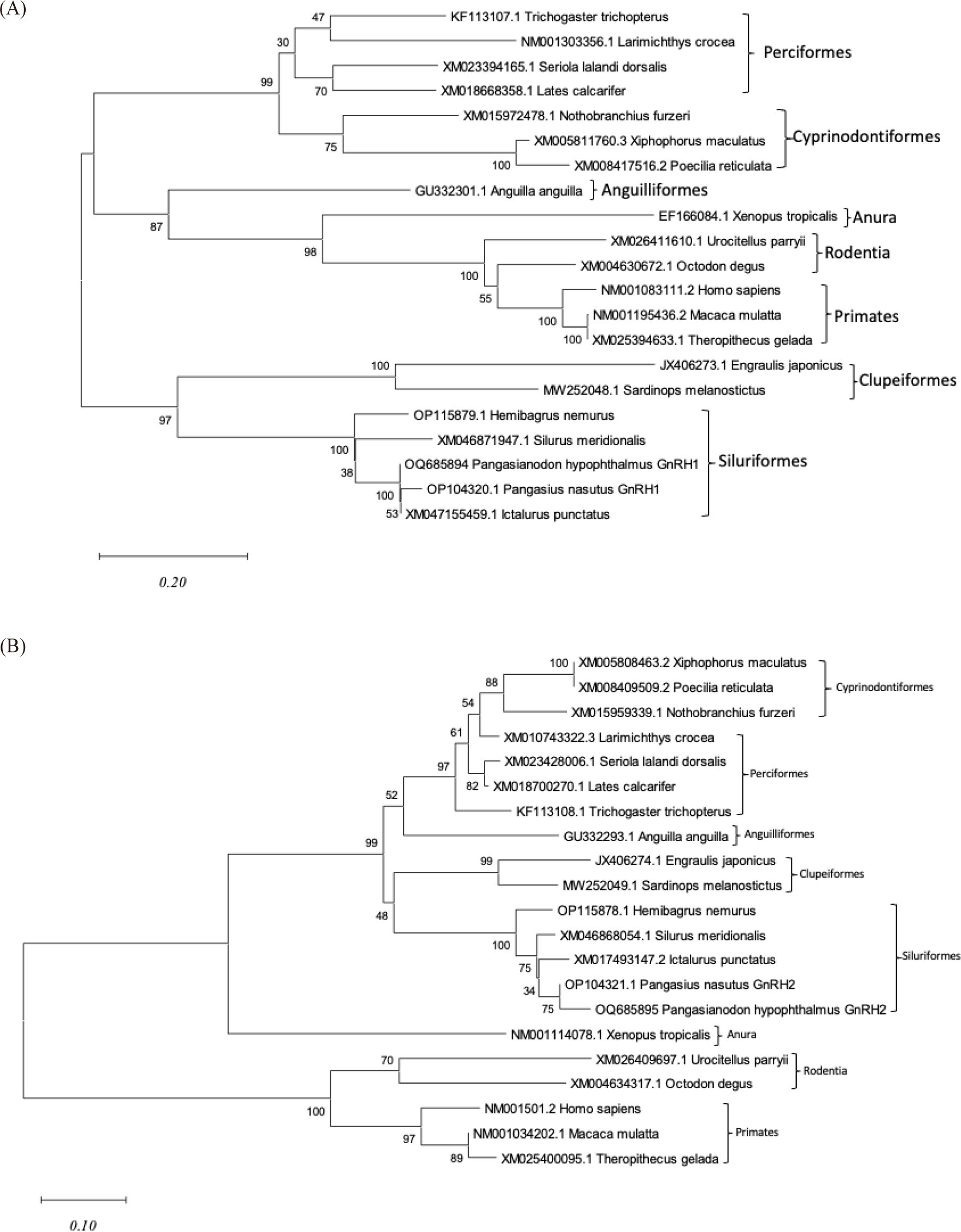
Since GnRH2 is highly conserved among the GnRH2 decapeptide across the vertebrates, the phylogenetic tree is formed according to their conserved region in the GAP region. Referring to Fig. 5B, the phylogenetic tree of GnRH2 segregated into two clades. The result of the phylogenetic tree for GnRH2 was in line with the result of multiple alignments (Fig. 4B). The first clade supported by a high bootstrap value (100%) consist of siluriformes, clupeiformes, anguilliformes, perciformes, cyprinodontiformes, and anura, having a fairly conserved GAP sequence peptide. Meanwhile, another clustered with a bootstrap value of 100% consists of rodentia and primates, which have highly variable GAP peptide sequences.
Discussion
This is the first report of characterization of the full-length cDNA gonadotropin-releasing hormone (GnRH) genes, GnRH1 and GnRH2, from P. hypophthalmus and P. nasutus. The PCR product has clearly shown the efficiency of the designed primers to amplify the targeted genes (Fig. 1). Study from Gaillard et al. (2018) reported the expression of the GnRH genes is exclusively detected in brain. The GnRH1 mRNA appeared particularly abundant in the diencephalon and at slightly lower levels in the telencephalon and mesencephalon, while GnRH2 mRNA found to be predominant in the mesencephalon and present in the diencephalon. However, GnRH3 is not detected in P. hypophthalmus and P. nasutus. Previous research revealed that fish from the siluriformes order only have two types of GnRH, which are chicken GnRH-II (cGnRH-II) and catfish GnRH (cfGnRH) (Klausen et al., 2001; Qin et al., 2022). Presence of these two putative GnRH peptides is consistent with a previous study performed on the closely related species, C. gariepinus (African catfish) showing the presence of two immunoreactive GnRH forms, identified as catfish GnRH, cfGnRH (GnRH1) and cGnRH-II (GnRH2) (Kumar et al., 2005). These findings are also supported by Muñoz-Cueto et al. (2020) eels and catfish appear to have lost the GnRH3 gene, however GnRH1-expressing neurons have been discovered in the olfactory bulbs and terminal nerve, which appear to substitute for the function of the GnRH3.
Generally, GnRH is all encoded as a preprohormone with a typical structure consisting of an UTR signal peptide, GnRH decapeptide (which is the bioactive peptide), three amino acid cleavage site (GKR), and the GAP (Chaube et al., 2019; Gaillard et al., 2018; Lumayno et al., 2022; Qin et al., 2022; Whitlock et al., 2019). The cleavage site GKR (Gly-Lys-Arg) sequence separates the hormone moiety from the GAP (Chaube et al., 2019). The majority of vertebrates GnRH have 10 amino acids, and the consensus sequence is pyro-Glu1-His2-Trp3-Ser4, Gly6, and Pro9-Gly10-amide (Sakai et al., 2020). The GnRH1 decapeptide is highly conserved at positions 1–4, 6, 9, and 10, five from the 10 amino acids being invariant, whereas the other two positions exhibit conservative modifications (Sower et al., 1997; Zmora et al., 2002).
The GnRH1 decapeptide sequence (QHWSHGLNPG) of P. nasutus and P. hypophthalmus is highly conserved among the siluriformes order, where there are few differences at positions 5 and 8 in the decapeptide region in other vertebrates. Comparing the three types of GnRH isoforms, the GnRH1 peptide sequence appears to be far more variable; with GnRH1 amino acid sequences are conserved particularly pGlu-His-Trp-Ser at the N-terminus, and the C-terminal is fixed to Pro-Gly-NH2, which are required for GnRH1’s physiological function (Choi, 2018; Gaillard et al., 2018; Sower, 1997). Amino acid at position 8 is the most variable amino acid; where if an arginine residue was at the 8th position, it will improve the receptor binding (Choi, 2018; Flanagan & Manilall, 2017). Variations also spotted at positions 6, 5, and 7. The variety in amino acids is due to the fact that GnRH1 decapeptide exhibits species-specific structural diversification (Muñoz-Cueto et al., 2020).
Meanwhile, the GnRH2 decapeptide of P. nasutus and P. hypophthalmus contains a core decapeptide (QHWSHGWYPG) region that is highly conserved between teleost fish and other vertebrates. In this study, GnRH2 is well conserved in siluriformes, perciformes, cyprinodontiformes, clupeiformes, anguilliformes, anura, rodentia, and primates without any modifications in the sequence. Kavanaugh et al. (2008) mentioned that the GnRH2 peptide is entirely conserved in all other vertebrate species, with the exception of the lamprey. Although the GnRH decapeptide is evolutionary conserved, there is divergence in the signal peptide and GAP. Study from Andersen & Klungland (1993) revealed that GAP could contribute to the structure and stability of prepro-GnRH, as well GAP may contribute in the folding and processing of GnRH prohormone and the control of gonadotropin secretion (Mohammadzadeh et al., 2020; Qin et al., 2022; Wetsel & Srinivasan, 2002). Findings from this study are consistent with other studies showing that GAP coding sequences are highly varied even within closely related groups of animals, showed a unique feature of the GnRH preprohormone despite the relatively high conservation of the GnRH gene structure and peptide sequences (White et al., 1995; Whitlock et al., 2019).
The amino acid sequence alignment of P. hypophthalmus and P. nasutus are most closely related to the GnRH genes of H. nemurus, I. punctatus, and S. meridionalis, which is consistent with the results of amino acid identity comparison and phylogenetic tree analysis. By comparing the amino acid sequence encoded by GnRH genes of different species, it was revealed that the primary amino acid identity for GnRH1 and GnRH2 genes shared a high degree of homology (> 99%) with fish species, indicating that the conserved structure of decapeptide might play an important biological function between different species. Phylogenetic trees showing the evolutionary relationship of GnRH1 and GnRH2 genes with other varieties of vertebrate taxa was generated using the neighbour-joining method. The phylogenetic analysis revealed that the vertebrate GnRH proteins clustered into clades according to their order: cyprinodontiformes, perciformes, anguilliformes, clupeiformes, siluriformes, anura, primates, and rodentia, displayed a close relationship among similar GnRH in different fish species. The constructed phylogenetic tree in this study correlated with previous studies (Chaube et al., 2019), which found that the GnRH2 protein of the catfish Heteropneustes fossilis was aligned closely with C. gariepinus GnRH2 and clustered with the GnRH2 type in the Ostariophysan subclade, distinct from the Acanthopterygian subclade, of teleosts. Phylogenetic analysis revealed that the vertebrate formed cluster in distinct clades and are grouped according to their GnRH decapeptide and GnRH-associated peptide, revealing a close relationship among similar GnRH decapeptide forms in different vertebrate species.
Conclusion
In summary, we successfully cloned and characterized the full-length cDNA sequences of the GnRH genes from P. hypophthalmus and P. nasutus. This study could generate the gene-specific species database of GnRH genes for commercially important catfish species, P. hypophthalmus and P. nasutus. Also, the findings should provide a basis for future reference for defining peptide functions, localization, and regulatory mechanisms of GnRH genes, as well as the development of recombinant GnRH hormone to be used for artificial breeding in the aquaculture sector.








