Introduction
In the seafood industry in Vietnam, shrimp accounts for approximately 40% of the total export turnover, bringing in nearly $ 3.4 billion in 2022. Because of this sector’s rapid growth and development, aquaculture wastewater in Vietnam is frequently dumped directly into natural water bodies, negatively affecting the aquaculture environment, biodiversity, and human habitats (Đôn, 2014). In addition, about 30% of nitrogen and 10% of phosphate (PO43–) from feed escape into the aquatic environment (Boyd, 2011). The abundant phosphate availability in a natural water environment is a typical reason for the dangerous eutrophication phenomenon (Yang et al., 2010). Therefore, PO43–-removal in forming polyphosphate (poly-P) via microbial activities have recently increased interest because of its low costs, convenient operation, and environmental friendliness (Chu, 2006; Ngô, 2012). More specially, the application of poly-P as a reusable form for developing raw materials in the fertilizer industry or an alternative phosphate source in carp diet in recent approaches has trigged focus on phosphorus recovering solutions from many wastewater systems through the bacterial poly-P accumulation mechanisms (Hirota et al., 2010; Yoon et al., 2016). Thus, screening microbial strains that can accumulate phosphate PO43–is not only an essential aspect of treating phosphorus compounds in the aquaculture wastewater system (Flowers et al., 2013; Oehmen et al., 2007), but also providing Poly-P for the fertilizer industry or biominerals from refined phosphate-accumulating organisms (PAOs) biomasses (Simoes et al., 2020; Yuan et al., 2012).
In water, PO43– compounds are the simplest forms of accumulation poly-P into the microorganism cells. For example, the PO43–-accumulating capacities of four PAOs, namely Pseudomonas, Enterobacteriaceae, Alcaligenes, and Streptococcus have recently been reported (Bao et al., 2007). However, their PO43–-accumulating traits are still the subject of debate because Acinetobacter or Accumulibacter phosphatis shows PO43–-accumulation but has never been cultured successfully in vitro experiments (Günther et al., 2009), in contrast to Microlunatus phosphovorus has even cultures in pure isolation but cannot determine precisely the PO43–-accumulating capacities into the cells (Mao et al., 2015). Furthermore, applying PO43– uptake assays can more conveniently confirm the PO43–-accumulating capacity upon the remaining phosphate content in cultural media than other methods (Aravind et al., 2015; Chaudhry & Nautiyal, 2011). Therefore, evaluating the performance of PO43–-removal systems is not always dependent on the proliferation of PAOs but also requires the successful in vitro cultivation of the PAOs (Tarayre et al., 2016).
Until now, intracellular poly-P of PAOs has also been detected as metachromatic granules, with an average size from 0.25 to 1.33 µM, which are not visible unless stained. Each PAO strain can accumulate an average of 100 granules in its cell (Günther et al., 2009; Serafim et al., 2002). The most common technique used to verify the presence of intracellular poly-P is the poly-P granules stained with chemicals or fluorescent dyes before observing them under a light or fluorescence microscope (Serafim et al., 2002; Tarayre et al., 2016). Recently, 4’,6-diamidino-2-phenylindole dihydrochloride (DAPI) dye with an optimal concentration of 10 µg/mL of DAPI can confirm the presence of PAO in sludge samples through poly-P granules with a green-yellow fluorescence, whereas the DNA shows blue fluorescence (Günther et al., 2009; Klauth et al., 2006; Terashima et al., 2020). Besides, although electron microscopy is a powerful technique that allows the visualization of the cellular location of poly-P, this method cannot confirm the dynamics in the linkages of poly-P granules with other compartments inside the bacterial cell (Lima et al., 2003; Watanabe et al., 2021). Although the mechanisms of poly-P aggregation have been proposed (Günther et al., 2009; Hupfer et al., 2008; Simoes et al., 2020; Serafim et al., 2002; Terashima et al., 2020; Watanabe et al., 2021; Ye et al., 2015) however, the processes involved in the formation of the poly-P in the PAO cell are still largely unknown.
Following the argument mentioned above, the diverse exist of microbial strains that can accumulate PO43– in an extensive shrimp pond in La Gi district, Binh Thuan Province in Vietnam at a nursing period of 4 to 5 months accrued significant amounts of phosphorus makes an ideal survey for studying the process and the accumulating properties of poly-P chains of PAOs. Our study successfully isolated four PAOs from eight colonies with a visibly resolved PO43–zone on the selective acetate mineral mediums (AMM) agar plate from water and sludge samples of the shrimp pond. The PO43–-accumulating capacities of four native PAOs (A1, A2, A3, and A5) and pH changes from the growth of PAOs in an AMM medium with and without five salt concentrations (from 0‰ to 30‰) at each differential cultural period were investigated detailly. The high homologous identities of nucleotide sequences of the polymerase chain reaction (PCR) products with 16S rDNA deposited genes in GenBank determined strain A2 was a Rhodotorula sp. In contrast, strain A1 was Acinetobacter guillouiae, A3 was Acinetobacter lwoffii, and A5 was Lactococcus raffinolactic. Remarkably, fluctuations in the fluorescence properties between DNA and poly-P in the PAO cells during culturing comprehensively discovered the assembly of poly-P chains in L. raffinolactic (A5) and the generation of poly-P granules underlying PO43–-accumulating in other trains, which can keep our knowledge about the poly-P accumulating process in PAO cells.
Materials and Methods
Isolating PAO strains were performed at an extensive shrimp pond with a pond area of 800–1,000 m2 in La Gi district, Binh Thuan Province in Vietnam, after the breeding shrimp nursing period of 4 to 5 months. In October 2020, water and sludge samples from this pond were collected to screen PAOs. After collection, these samples were stored at 4°C–10°C and transported to the laboratory. One gram of the sludge sample and 50 mL of the water sample were placed into a beaker and gently mixed using a glass float to obtain a homogeneous representative sample. Subsequently, 1 mL of the representative sample was placed into a test tube containing 9 mL of 0.9% NaCl and mixed to obtain the 10–1 dilution. Five different ranges of bacterial suspensions from 10−1 to 10−5, 50 µL of each diluted concentration, were plotted on plates containing selective AMM in triplicates and then incubated at a temperature of 25°C for 3–5 days in an incubator shaker at 30,000×g to isolate PAOs (Bao et al., 2007). After incubation, colonies forming clear surrounding halos were identified as PAOs (Nautiyal, 1999). Single colonies were sub-cultured on AMM to obtain pure colonies, which were examined microscopically and morphologically. Each colony differs in shape, color, and size, identified via Gram staining. Subsequently, potential PAOs were sub-cultured in a selective medium and spiked with 20% glycerol for storage at –70°C.
The antagonistic activity of PAOs was assessed by the streak line method on LB agar under aerobic conditions. First, 10 µL of each inoculated PAO strain (24 hours at 25°C) was streaked along a straight line on the LB agar plate with a transfer rod and dried. Next, the three remaining strains were streaked at right angles of the straight line before incubating the agar plate at 37°C for 24 or 48 hours. The antagonistic ability of all PAOs was determined by the inhibition zone (in millimeters) that appeared on the LB agar, ranked as high (> 25 mm), intermediate (13–25 mm), and low (0–12 mm) (Hütt et al., 2006; Nguyễn & Phan, 2014).
To determine the effect of the salt concentration on the growth of PAOs, the salt tolerance and thermal tolerance capacity of the isolates were checked by cultivating the strains in LB medium supplemented with NaCl at concentrations of 0‰, 5‰, 10‰, 20‰, and 30‰. Incubation time was 12 to 48 hours at 25°C. During the culture period, at surveying time points, 1 mL of the inoculated medium was aspirated and photometrically measured at 600 nm to investigate the impact of the salt concentration on growth was determined. All experiments were performed in triplicate.
The PO43–-solubilization is a crucial trait of bacteria and is routinely measured to assess PO43–-accumulation of PAOs. Their growth in AMM was also investigated regarding the PO43–-accumulation of PAOs. The composition of the AMM medium was as follows (for 1 L): 10 g C6H12O6, 5 g Ca3(PO4)2, 5 g MgCl2 · 6H2O, 0.25 g MgSO4 · 7H2O, 0.2 g KCl, 0.1 g (NH4)2SO4, 15 g agar. The PO43–-accumulation was investigated by cultivating 500 µL of isolated PAO stocks in test tubes containing 7 mL of AMM for 20 hours at 25°C and 1.035×g. After that, we transferred 1 mL of the culture medium to a conical flask containing 100 mL of AMM supplemented with 5% K2HPO4. The continuous culture’s cultivation periods were 24, 48, 54, 72, 96, and 120 hours. For each period, the total volume was centrifuged at 7.168×g for 10 min to collect the supernatant. Subsequently, 750 mM of Ames solution was sequentially added to 250 µL of the supernatant, followed by incubation at 37°C for 20 min for color development. The Ames solution consisted of 0.36% (w/v) NH4)6Mo7O24. 4H2O in 1 N H2SO4 plus 1.42% ascorbic acid and was prepared immediately before the assay. Each assay was performed in triplicate, and each isolate’s average value was recorded as the PO43–-accumulation capacity. Pure K2HPO4 was used as the standard reagent. The PO43–-accumulation capacity of PAOs was determined based on the residual PO43–-content in the medium that reacted with ammonium molybdate to form a complex in an acidic environment. This complex is reduced sequentially by ascorbic acid to form a blue complex with an absorbance of 820 nm (Ames & Dubin, 1960; Barrows et al., 1985; Worsfold et al., 2005), which can be used to determine PO43–-accumulation capacity of each PAO. The residual PO43–-concentration in the medium was measured using a spectrophotometer (Genesys 10S UV-Vis, Thermo Scientific, Waltham, MA, USA) at 820 nm. The PO43–-accumulation was calculated from the changes in absorbance, applying Equation (1):
where C0 represents the initial concentration, and Ct indicates the concentration at time t.
As described above, 500 µL of isolated PAO stocks in test tubes containing 7 mL of AMM at 25°C with shaking at 1.035×g for 24 to 48 hours, depending on each strain. After incubation, 1 mL of the culture was transferred to a conical flask containing 100 mL of AMM supplemented with 5% K2HPO4 and NaCl at 5‰, 10‰, 20‰, and 30‰, followed by incubation at 25°C under shaking at 1.035×g. At different time points (24, 48, 72, 96, and 120 hours), samples were collected by centrifugation at 7.168×g for 10 min to determine the concentration of PO43– via spectrometry at 820 nm. The PO43–-accumulation ability under different salt concentrations was calculated using Equation (1); each experiment was performed in triplicate.
The genomic DNA of the four microbial strains was extracted using the 95°C heat shock method. The extracted DNA was then used as template for PCR amplification with two pairs of primers specific for the 16S rDNA gene fragment. The forward primer was 27F (5’-GAGTTTGATCCTGGCTCAG-3’), and the reverse primer was 1492R (5’-AAGGAGGTGATCCAACC-3’). The primer for the ITS1 region of the forward sequence was ITS-F (5’-TCCGTAGGTGAACCTGCG-3’), and the reverse primer was ITS-R (5’-GCTGCGTTCTTCATCGATGC-3’) (Bao et al., 2007; Mokhtari et al., 2011; Zhang et al., 2015). We used 5 µL of template mixture for the PCR reaction. The PCR products were approximately 1.500 bp and 470 bp-long gene fragments of the 16S rDNA and ITS1 regions, respectively, and processed via 1.5% agarose electrophoresis. Subsequently, the harvested PCR amplicons were sequenced, and the final sequences were submitted to the GenBank database (National Center for Biotechnology Information, NCBI) to obtain the corresponding accession numbers.
The BLAST tool was used for phylogenetic analysis to search for homologous sequences previously published by other authors and stored in the NCBI. A phylogenetic tree was constructed using the MEGA 7.0 software, employing the neighbor-joining DNA distance algorithm with a bootstrap of 1.000. Then, comparing the position differences between species pairs using the software and building a phylogenetic tree based on the neighbor-joining method, the index at the base of the branches had a bootstrap value of 1.000 resampling times (%). The confidence level assessed according to the NJ bootstrap was below the high confidence level of > 85%; the average confidence level was 65%–85%, and the low confidence level was 65%, according to the MEGA 7.0 software.
Firstly, ten µg/mL of DAPI (Sigma-Aldrich, St. Louis, MO, USA) (Tijssen et al., 1982) was used for staining 400 µmol/g dry weight cell streaks containing poly-P and DNA particles. The accumulation of intracellular poly-P inside the cell was visualized by microscopic observation (Klauth et al., 2006; Streichan et al., 1990). The bright yellow-fluorescent response of poly-P is a vital indicator for the discrimination of the blue-fluorescent DNA. First, 1 mL of the culture solution of the four PAO strains was grown proliferatively for five different culture periods and collected in a 1.5 mL Eppendorf tube, followed by centrifugation at 3.200×g for 5 min. The cell pellet was washed twice with 200 µL of Mcllvaine’s buffer (two parts stock solution A: BaCl2 · 2H2O, 5 mM NiCl2 · 6H2O and eight parts stock solution B: 0.4 M Na2HPO4/NaH2PO4, 150 mM NaCl, pH 7.2) to remove any interfering substances (cellular debris or organic matter), resuspended in 4% formaldehyde, and incubated for 30 min at 25°C. Next, the cell pellet was rewashed with 200 µL of Mcllvaine’s buffer before being stained with DAPI. Subsequently, we dissolved 2 mg DAPI in 1 mL Mcllvaine’s buffer to prepare a stock solution with a concentration of 2 mg/mL and, subsequently, a working stock solution with a concentration of 10 µg/mL for the following staining process (Serafim et al., 2002; Terashima et al., 2020; Watanabe et al., 2021). Next, to stain double-stranded DNA and poly-P with DAPI, 200 µL of the working solution was added to the treated cell pellet, followed by incubation for an additional 30 min at room temperature (25°C). The stained samples were centrifuged at 1.080×g for 5 min, and the pellets were resuspended in 20 µL phosphate-buffered saline (PBS, pH 7.2, Sigma-Aldrich). Subsequently, 2 µL of the stained mixture was subjected to fluorescence microscopy at the 450–650 nm range, with excitation at 360 nm (Serafim et al., 2002). The treatments were performed in triplicate.
Microbial growth and the pH values in the mixed liquid were monitored by an optical density (OD) analyzer and a pH analyzer, respectively. The OD concentration was determined by using a UV-Vis instrument at a wavelength of 600 nm. All statistical analyses were performed with GraphPad Prism 8.3.0. The data were subjected to a two-way analysis of variance (ANOVA). Specific differences between individual means from a set of standards were detected using Tukey’s test, and differences at p < 0.05 were considered significant.
Results
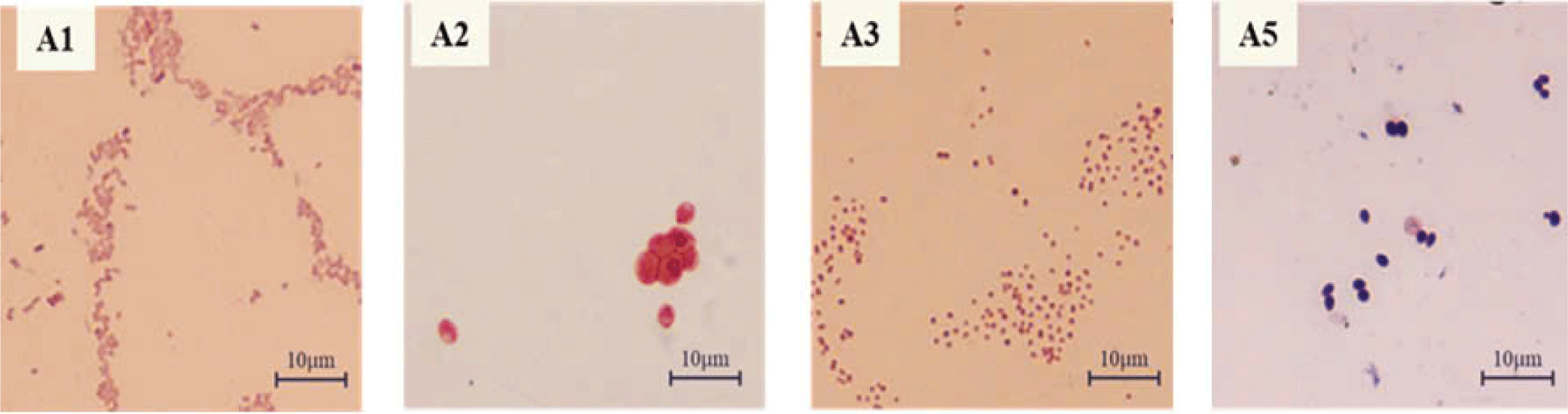
The 100 µL of 10–5 diluted solution samples from shrimp ponds on sterilized AMM-selected isolated thirty-two colonies with various morphological characteristics, mainly milky white, milky white, yellow, or pink circular colonies. After that, eight colonies with a visible zone on the selective AMM agar plate were isolated as potential PAOs from the initially screened 32 strains with the ability to solubilize PO43– (Supplementary Fig. 1S). Four of the colonies exhibited rates of PO43–-solubilization over 50% in 48 hours and were selected as A1, A2, A3, and A5. The selected strains were then subjected to Gram staining to observe their morphological characteristics. As shown in Table 1, strains A1, A3, and A5 appeared as white colonies with a round shape and entire edges, whereas strain A2 had an oval shape and was pink. Strains A1, A3, and A5 were Gram-negative bacteria, whereas strain A2 was a yeast (Fig. 1). Subsequently, the strains were preserved in a freezer at –70°C for the identification experiments.
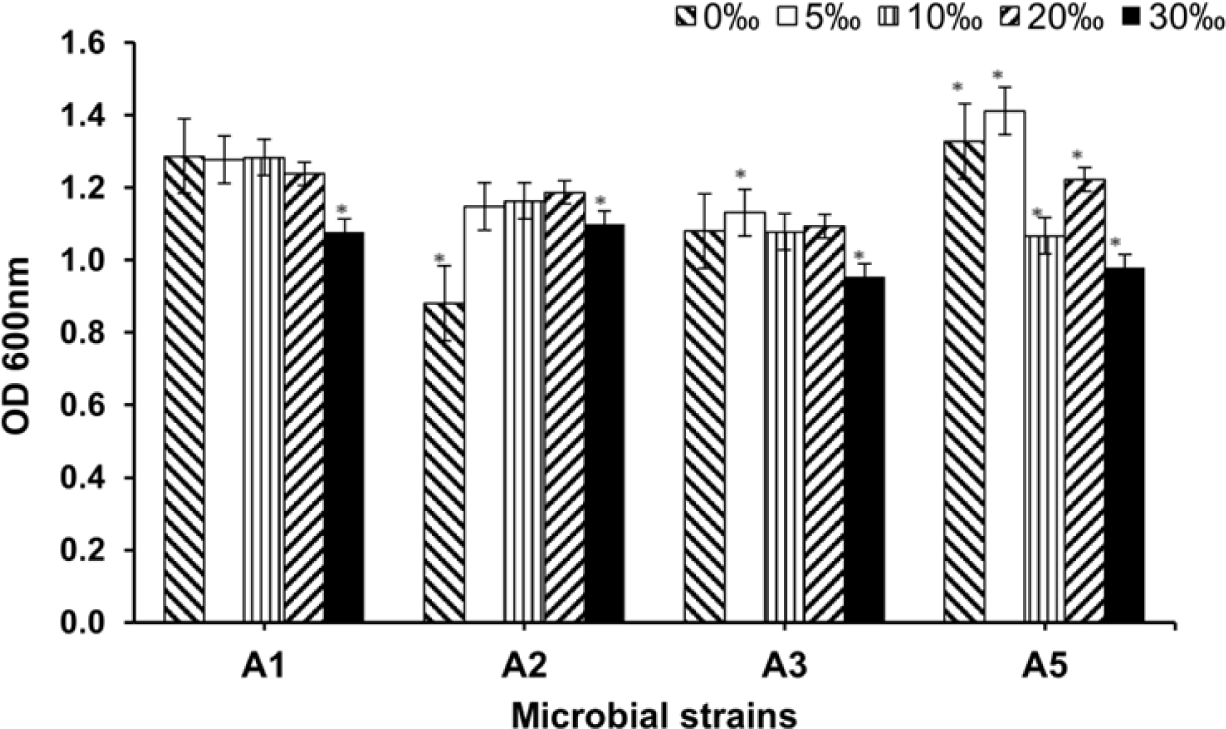
The absorbance of bacteria at 600 nm was used to estimate bacterial growth at the four NaCl concentrations. As shown in Fig. 2, the growth of A1, A2, and A3 did not differ significantly among the different salt concentrations (p > 0.05), whereas strain A5 was affected considerably by NaCl, irrespective of the concentration. For A1, growth was most pronounced (OD600nm of 1.3A), with negligible fluctuations. In contrast, the growth of strains A2, A3, and A5 changed slightly depending on the NaCl concentration. However, all strains showed decreased growth at a NaCl concentration of 30‰. At the highest salt concentration, strains A1 and A2 were more stable, with an OD600nm around 1.1A, compared to the other strains, with a value of approximately 1.0A. The above differences may be explained by the different environments that interfered with the growth abilities of the strains. These results indicate that the different strains differed in their ability to adapt to a salt concentration of 30‰.
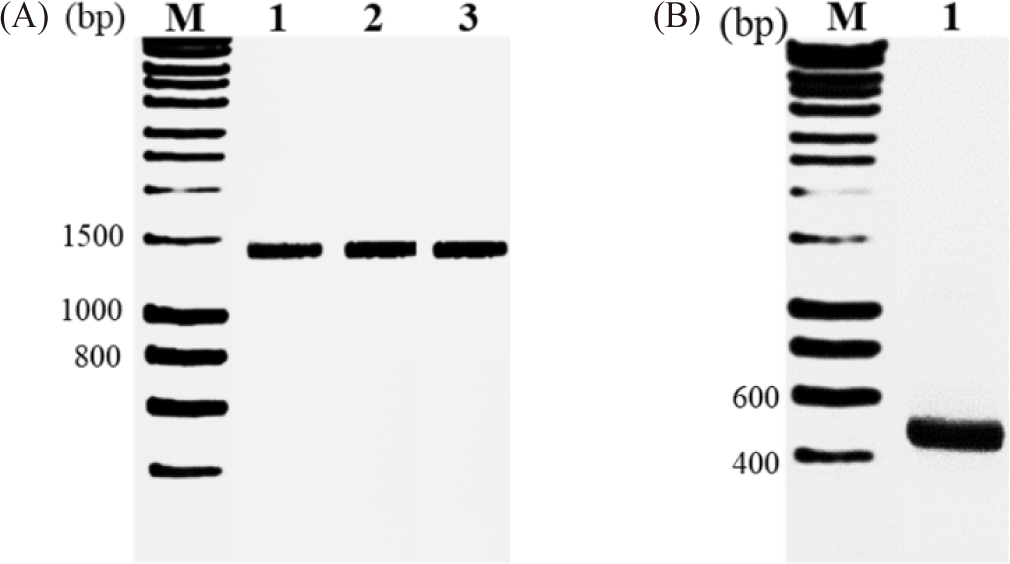
According to previously described methods, the 16S rDNA gene sequencing was carried out for pure representative cultures of the four isolates. Hence, with the presence of PCR products of 1.500 bp for 16S rDNA primers and 470 bp for ITS1 primers on 1.5% agarose gel, each primer pair gave amplified products of the expected size (Fig. 3). These findings agreed with the results reported by Mokhtari et al. (2011). The amplified products were sequenced, and the 16S rDNA gene sequences of three strains, A1, A3, and A5, obtained 1.489 bp with stable signals. After submitting the 16S rDNA to the GenBank database and comparing it against known gene sequences using the BLAST tool, the resulting sequences matched those of their respective type strains. Based on the aligned results, the DNA of strain A1 was 100% identical to that of A. guillouiae MT310710.1. The DNA of strain A3 showed 99.71% identity with that of A. lwoffii MH267196.1, and the DNA of strain A5 was 100% identical to that of L. raffinolactic HM058735.1. Finally, an aligned result of the ITS1 sequence region of the A2 strain of 450 bp stable nucleotides against known gene sequences using the BLASTn tool in NCBI showed a 100% similarity in nucleotide sequences with those of the genus Rhodotorula sp. MN268779.1.
The phylogenetic tree was built from the 16S rDNA sequences of strains A1, A3, and A5, compared to the NCBI. The diagram of the phylogenetic tree in Supplementary Fig. 2S shows that strains A1 and A3 belong to the genus in a subgroup of the phylogenetic tree, with sequence similarity among species of the genus Acinetobacter, based on the NCBI database. Strain A1 showed no difference in base number compared with A. guillotine MT310710.1 with a bootstrap index of 100. Strain A3 differed from A. lwoffii MH267196.1 by a bootstrap index of 74. Unlike the above two strains, A5 was identified as L. raffinolactic HM058735.1 by a difference in the number of base sequences and a bootstrap index of 71 (Supplementary Fig. 3S). Similarly, the diagram in Supplementary Fig. 2S shows that strain A2 belongs to the genus Rhodotorula sp. MN268779.1, which has a base number difference in the sequence; the bootstrap index was 93.
Compared to the NCBI, the phylogenetic tree was built from the 16S rDNA sequences of strains A1, A3, and A5. The diagram of the phylogenetic tree in Supplementary Fig. 2S shows that strains A1 and A3 belong to the genus in a subgroup of the phylogenetic tree, with sequence similarity among species of the genus Acinetobacter, based on the NCBI database. Strain A1 showed no difference in base number compared with A. guillotine MT310710.1 with a bootstrap index of 100. Strain A3 differed from A. lwoffii MH267196.1 by a bootstrap index of 74. Unlike the above two strains, A5 was identified as L. raffinolactic HM058735.1 by a difference in the number of base sequences and a bootstrap index 71 (Supplementary Fig. 3S). Similarly, the diagram in Supplementary Fig. 2S shows that strain A2 belongs to the genus Rhodotorula sp. MN268779.1, which has a base number difference in the sequence; the bootstrap index was 93.
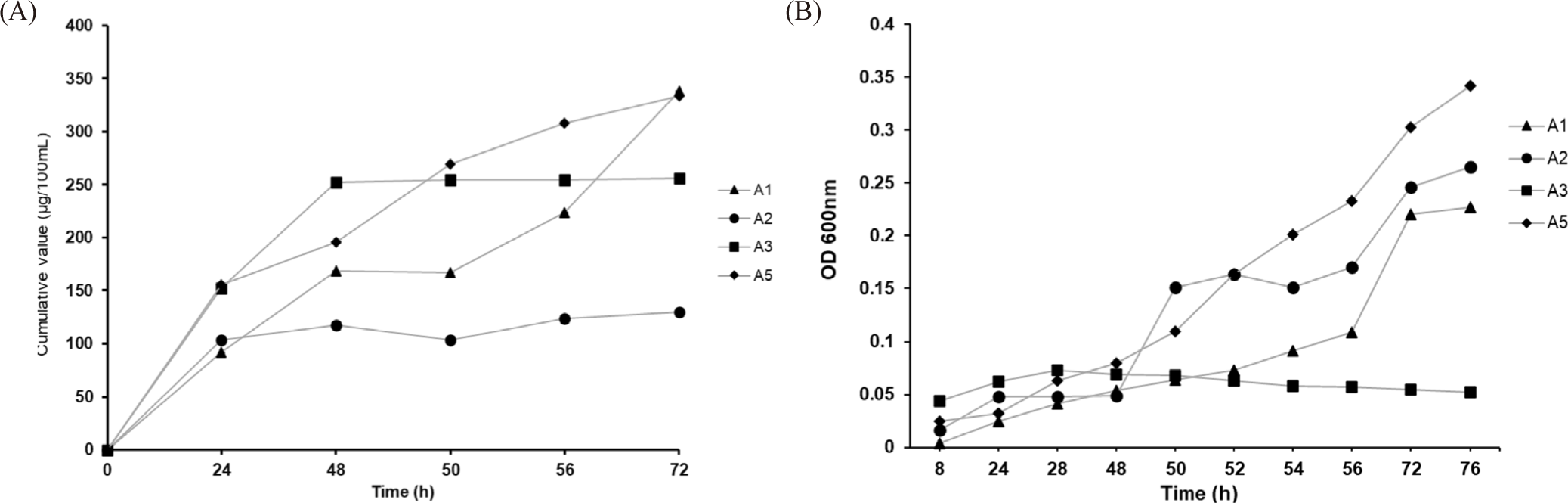
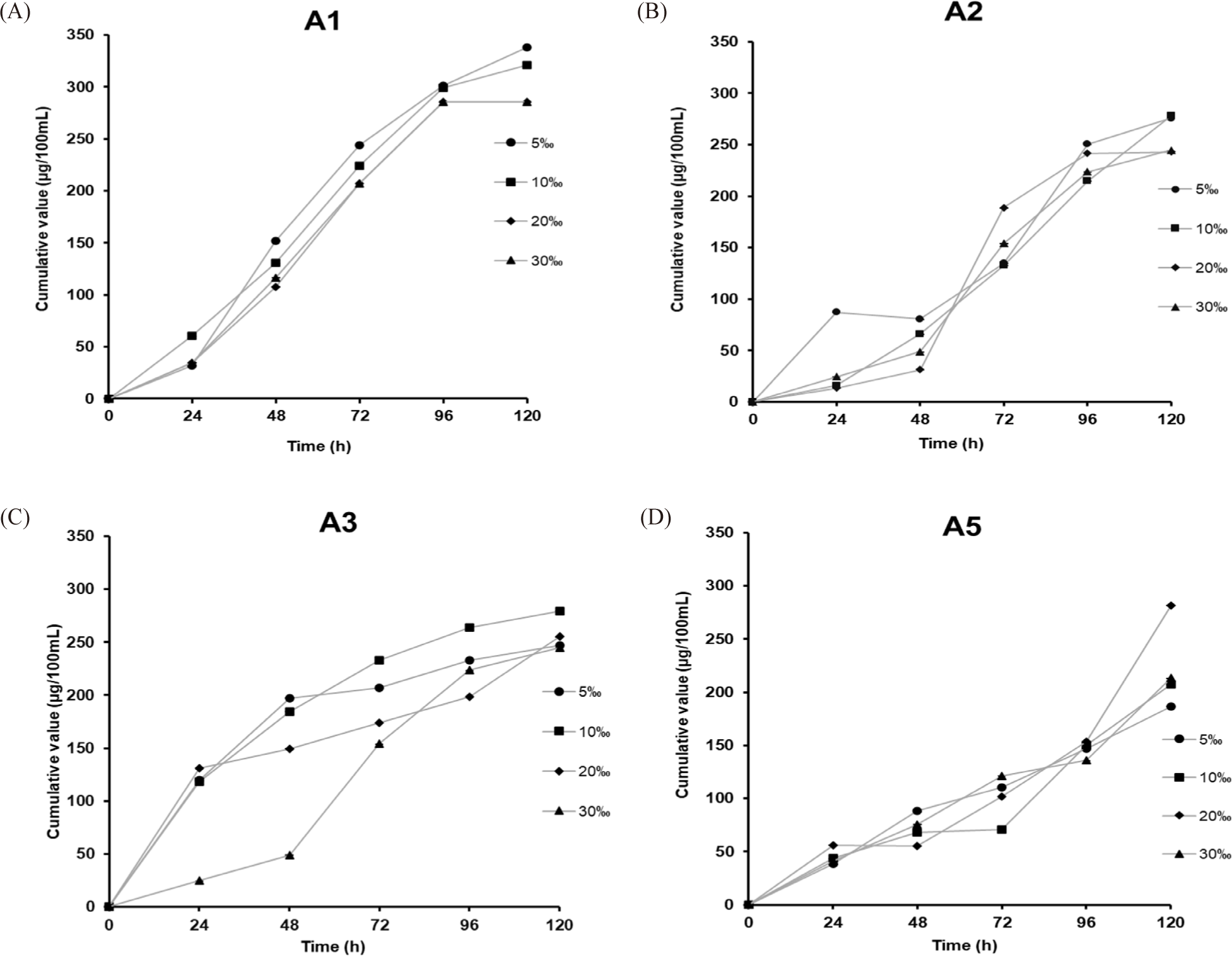
After the inoculation periods, the residual PO43–-content in the AMM was the PO43–-accumulation from the Ca3(PO4)2 component of each PAO strain. Changes in cumulative values correlating to the PO43–-accumulating ability of each PAO strain are shown in Fig. 4 and Table 2. Strains A1 and A5 showed gradually increasing PO43–-accumulating capacities. In contrast, those of the remaining strains (A3 and A5) only described the fast acquisition in the initial 48 hours and the next time exhibited relatively constant, and finally did not involve any changes during later periods. The relative PO43–-content in the AMM for each inoculation period, as shown in Table 2, revealed that strains A1 and A5 showed the highest respective accumulation, with 338.44 and 333.63 μg/100 mL after 72 hours. In contrast, strain A3 only accumulated 256.42 μg/100 mL. The lowest value was found for strain A2 with 130.19 μg/100 mL. Because four PAOs lived and the PO43–-content in the initial AMM solutions was not the same, it indicated the different accumulation capabilities of the PAOs (p < 0.05).
All data has presented as mean ± SE (n = 3) statistical differences (p < 0.05) with Multiple’s ranges test. The initial values provided differences of each inoculate period. The second value provided differences of each NaCl content.
Data from Bao et al. (2007).
The growth behavior and pH values in the selective AMM over 76 inoculation hours are shown in Fig. 4B. In general, the growth rate of the A3 strain was the lowest, and the difference in the growth of all four PAOs can be understood as a response to the different nutrient levels. Besides, the pH values measured during the inoculation periods fluctuated around 7.5 to 8. As seen in both Fig. 4A and 4B, although the PO43–-accumulation efficiency of the A1 strain was not higher than that of the A5 strain, both showed the same growth behavior. Based on the initial PO43–-accumulation in AMM, we further investigated the impact of salt concentration on PO43–-collection of all PAOs.
Fig. 5 shows the PO43–-accumulation of the four PAO strains in AMM medium with a PO43–-content of 340 μg/100 mL supplemented with NaCl at five different concentrations (5‰, 10‰, 20‰, and 50‰). The fluctuations in the remaining PO43–-content showed a slight impact of the NaCl concentration on PO43–-accumulation for all four PAOs (p < 0.05). As shown in Table 2, strain A1 led the best performance, with efficiencies of 338.40 and 321.29 μg/100 mL at salt concentrations of 5‰–10‰ and a lower cumulative efficiency of 285.55 μg/100 mL at salt concentrations of 20‰–30‰ after 120 hours of culturing (p < 0.05). We observed independent variations in PO43–-accumulation for A3 and A5; the maximum collection was found for A5, with 281.65 μg/100 mL at a NaCl content of 20‰, followed by that for A3 with 279.70 μg/100 mL at a NaCl content of 10‰. Except for the A2 strain, adding NaCl content from 5‰–30‰ enhanced its PO43–-accumulation, resulting in an accumulation efficiency of 246.80–277.92 μg/100 mL after 120 hours of cultivation. Without NaCl addition, the maximum accumulation efficiency was 130.19 μg/100 mL after 72 hours of cultivation. These findings show that the microbial strains isolated here have been treated with the PO43–-content in an AMM medium with different salt concentrations.
Because intracellular poly-P cannot easily be observed without staining (Serafim et al., 2002), digital image analysis allowed us to visualize poly-P in PAO cells after 72 hours of cultivation in AMM under aerobic conditions (Mino et al., 1998). In a 2 µL sample containing each PAO stained with ten µg/mL DAPI, the type of poly-P forms inside the PAO cells could be distinguished by yellow-green fluorescence emission in contrast to the blue fluorescence of DNA (Fig. 6). After 72 hours of incubation, the yellow fluorescence of strains A1 and A5 was more intense than that of strains A2 and A3. DNA surrounded Poly-P in the cells of three PAO strains (A1, A3, and A5). In strains A1 and A3, poly-P comprised concentrated granules with yellow-green fluorescence, whereas A5 consisted of a poly-P chain. The location and contributions of poly-P and DNA particles in each PAO sample were observed in bright-filed images. The fluorescence signals of poly-P for strains A1 and A3 were like those reported for Corynebacterium glutamicum (Klauth et al., 2006).
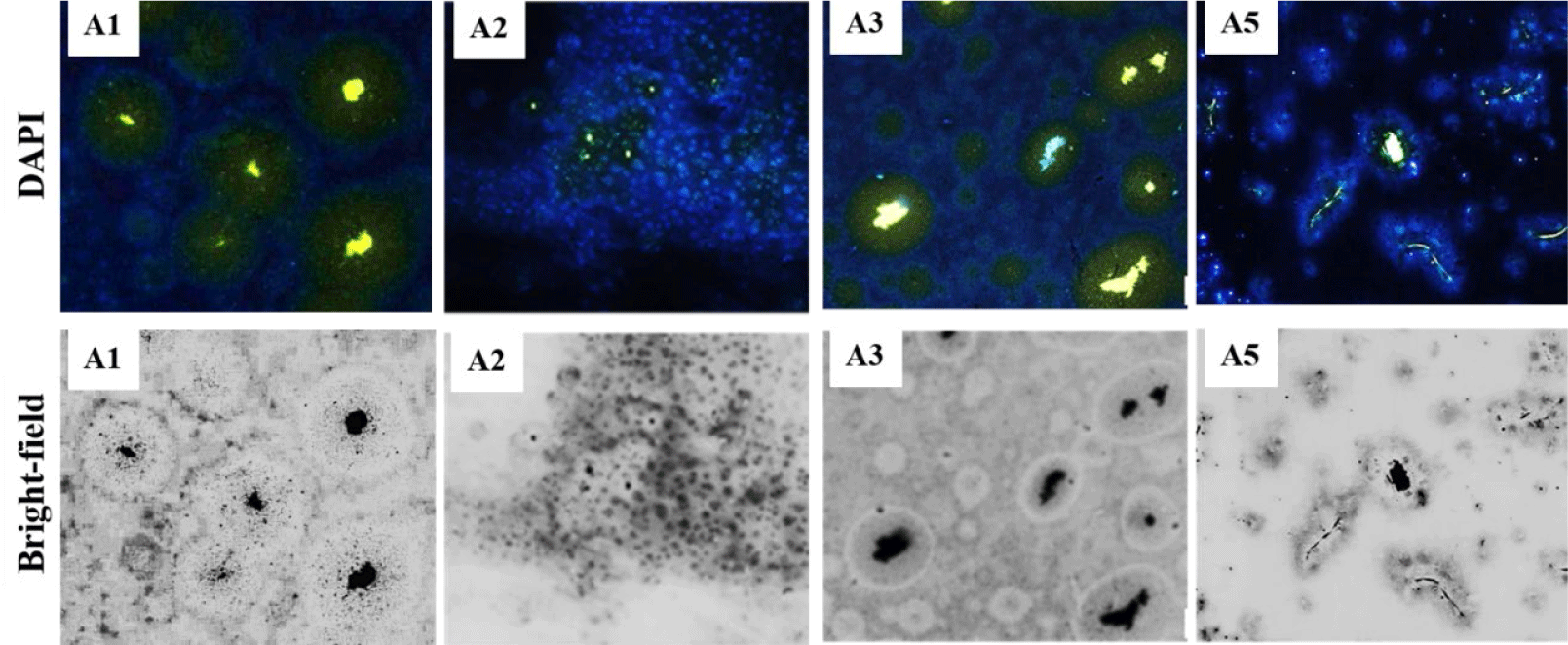
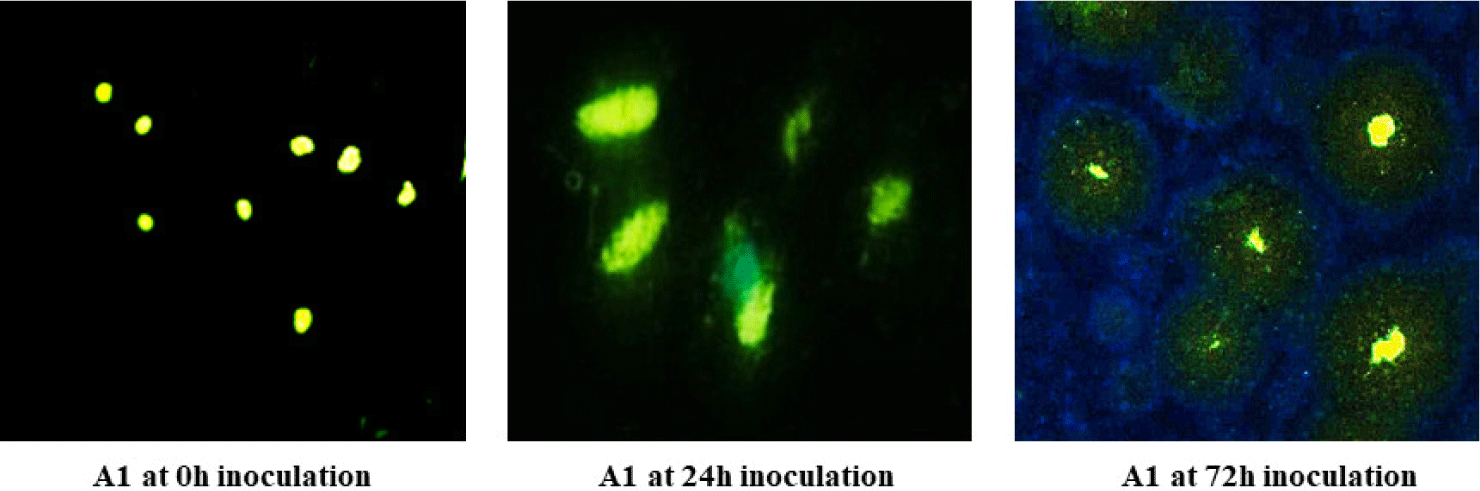
Fig. 7 shows the distribution of the yellow fluorescence signal of poly-P compared to that of the DNA for all PAOs at 0, 48, and 72 hours of incubation. The amount of poly-P fluorescence in all four PAOs substantially increased after 24 hours, followed by a rising population of poly-P in a sequential period to obtain the high intensity of the poly-P at 72 hours. Analysis of the poly-P content dynamics allowed us to determine the characteristics of the four PAOs. Remarkably, upon the profiles of the contribution of the yellow fluorescence contents in the A. guillotine cells (A1) strain, we can propose a PO43–-accumulation process beginning by slight yellow fluorescence responding to PO43–-content at the 0h inoculation time, followed by conjunction of yellow-green fluorescence of granule poly-P was linked to yellow-green fluorescence of DNA at 24 hours. At the end time of 72 hours, a phenomenon of concentrated poly-P granules with yellow fluorescence surrounded by DNA range was observed clearly inside A. guillotine cells (A1).
Discussion
PAOs, integral microorganisms in the natural phosphorus cycle, have recently attracted interest due to their effective PO43–-accumulation activity from aqueous solutions (Stokholm-Bjerregaard et al., 2017). This study successfully applied a commonly suitable approach to isolating PAOs involved in PO43–-accumulation ability by cultivating microorganisms in AMM. Their scientific names identified all four PAOs through phylogenetic analysis of conserved gene sequences performed for bacterial 16S rDNA and yeast ITS1 region. Among these, three bacteria, A1, A3, and A5, were found, including A. guillotine, A. lwoffii, and L. raffinolactic, while the fourth, A2, was identified as Rhodotorula sp.
With the lowest residual PO43–-content in the AMM with or without NaCl demonstrated, only A. guillotine got the highest PO43–-accumulation efficiencies during most inoculated periods compared to the three remaining strains. Next, adding NaCl content in the AMM revealed different trends in PO43–-accumulating capacities of all PAOs (p < 0.05). Although all PAOs might extend the inoculation periods to 120 hours for almost PO43–-accumulations in the NaCl-added AMM, A. guillotine showed a declined gradual impact of the PO43–-accumulation in AMM with ascending NaCl content (5‰ to 30‰), whereas the maximum PO43–-collections of L. raffinolactic with 281.65 μg/100 mL recognized at single condition of NaCl content of 20‰, followed that value was 279.70 μg/100 mL by A. lwoffii at a NaCl content of 10‰. One important thing is that only single yeast, such as Rhodotorula sp., first determined in this study, not only shows a higher capacity to accumulate PO43– in the AMM than that in A. lwoffii but also can more effectively accumulate the PO43–-content in the AMM containing NaCl at 5‰–30‰.
A comparative analysis of PO43–-accumulation efficiencies between four PAOS and four reference strains, Enterobacteriaceae, Streptococcus, Alcaligenes, and Pseudomonas, were summarized and displayed in Table 2. Except for Pseudomonas with the highest PO43–-accumulating efficiency (1.434 μg), three strains (Enterobacteriaceae, Streptococcus, Alcaligenes) with accumulation levels of 91, 174, and 224 μg (Bao et al., 2007) show the PO43–-accumulating efficiencies were lower than three PAO strains (A. guillouiae, Rhodotorula sp., and L. raffinolactic). Furthermore, the PO43–-accumulating capacities with different salinities combined maintain the pH of the AMM, around 6.5 to 7.0, providing evidence for the excellent adaptability of all four PAOs on survival conditions containing NaCl at 5‰–30‰. These results indicated impressing PO43–-accumulation behaviors of four PAOs compared to other referenced strains.
Poly-P has been constituted by an aggregation of PO43–, with an astounding number of activities performed in the cell. However, the most recent reports have yet to explain the PO43–-accumulating process of PAO in depth (Long et al., 2021). Previous studies reported poly-P content uptake in PAO cells during a batch cultural period from the large to the stationary phase with almost no fluctuations (Klauth et al., 2006; Zheng et al., 2020). On the other hand, although cytosolic poly-P presents a green fluorescence throughout the C. glutamicum cell (Klauth et al., 2006), this observation has not yet been displayed dynamically in connection to other components in the cell (Serafim et al., 2002). In addition, most studies on the observation of stored poly-P inside microorganisms, such as M. phosphovorus, Pseudomonas spp., and Paracoccus spp., detected and quantified in the granule conformations (Günther et al., 2009; Klauth et al., 2006; Terashima et al., 2020). Therefore, results from the present study proposed the first evidence of the assembly of poly-P chains in L. raffinolactic (A5) against the generation of poly-P granules in the three remaining strains. Remarkably, our findings comprehensively discovered the forming process of poly-P from fluctuations in the fluorescence signals of DNA and poly-P in A. guillotine cells through image analysis at three inoculation times (0, 24, and 72 hours); these results deepened our knowledge about the poly-P accumulating process in PAO cells.
Finally, although in explanation of the results comparing the distinct PO43–-accumulation abilities of four PAO strains in vitro investigation summarized here, the study has its limitations, such as cultural conditions and small scale that have yet to be fully simulated in natural environments. Hence, we proposed further research directions, such as testing the ability of PAO strain to treat wastewater on a larger scale and consequently in natural environments. In addition, studying the effects of other environmental factors (such as temperature and pH) on the process of poly-P accumulation is also available evidence to gain knowledge of the different properties of poly-P accumulation in each PAO strain.
Conclusion
The present research defined the PO43–-accumulation abilities from the 5 g Ca3(PO4)2 component in the AMM and the poly-P acquisition efficiency into PAO cells during 72 inoculate hours. These PO43–-accumulative processes have proven that all PAOs accumulated PO43– in the AMM into poly-P in the PAO cells. The best PO43–-accumulative efficiencies were A. guillotine (A1) and L. raffinolactic (A5), with 338.44 and 333.63 μg in 100 mL of AMM, respectively, after 72 hours under cultural AMM without adding NaCl. Notably, Rhodotorula sp. showed higher PO43–-accumulating capacities in higher NaCl-containing AMMs from 5‰–30‰. Although the NaCl contents (5%–30‰) slightly impacted inoculation time for the PO43–-accumulating capacities of PAOs, the activities of the four PAOs did not alter the pH of the AMM, around 6.5 to 7.0, providing evidence for the impressing PO43–-accumulating capacities of four PAO strains on different survival media conditions. With poly-P stained with DAPI, this study is the first report on the accumulation of PO43– to forming poly-P chains in L. raffinolactic cells against poly-P granules generated in cells of other strains. Based on distribution changes of poly-P at three inoculation periods (0, 24, and 72 hours), we can see the forming process of poly-P in A. guillotine cells. The PO43– removal in forming poly-P via microbial activities of four PAOs in this study are compelling features to support sustainable solutions for treating different PO43–-polluted water resources and further promises to provide valuable sources for harvesting -contained compounds that are applied in the fertilizer industry or biominerals from biomasses.







