Introduction
The order Cetartiodatyla, specifically the Infraorder Cetacea, encompasses a diverse group of marine mammals, includes whales, dolphins and porpoises. Among them, rorquals, classified under the family Balaenopteridae, are generally the largest, while dolphins are smaller and porpoises are the smallest in size (Wilson & Mittermeier, 2014). In addition to external morphological characteristics, one key distinguishing feature of whales is the presence of keratinaceous baleen plates, used for filter feeding, in contrast to the conical-shaped teeth of dolphins and spade-shaped teeth of porpoises (Jefferson et al., 1993). The family Balaenopteridae comprises nine species under the genus Balaenoptera and a single species under the genus Megaptera. These enigmatic marine mammals face several anthropogenic threats including direct exploitation through deck-mounted harpoon cannons, exposure to pollutants, and vessel strikes (Kemper et al., 2005). Consequently, live sightings of these animals are becoming increasingly rare (Sutaria et al., 2016). Nevertheless, occasionally, deceased cetaceans wash up on the Indian coastline, which extends for 7,516 km, encompassing nine coastal states, two union territories, and two island territories in India.
These stranded animal carcasses often provide invaluable insights into the natural history, ecology, and population genetics of these species (Bilgmann et al., 2011; Tucker et al., 2019). Moreover, records of marine mammal strandings along coastlines contribute relevant information for conservation efforts and offer insights into the status of marine ecosystems (Coombs et al., 2019). Furthermore, the long-term monitoring and detailed autopsy examinations of these stranded animals are crucial for determining the cause of death and guiding future conservation measures (Arbelo et al., 2013). However, identifying species from deteriorated biological samples in the maritime environment can be a challenging process.
To overcome the taxonomic challenges associated with species identification, molecular approaches have proven effective in whale species recognition and systematics research (Ross et al., 2003). The DNA sequences of the mitochondrial genome have provided new insights into cetacean origin and evolution (Arnason et al., 2004). Notably, two mitochondrial ribosomal RNA genes, as well as the protein-coding genes COI and Cytb, have been demonstrated to effectively discriminate major cetacean groups (Milinkovitch et al., 1994; Sholl et al., 2008; Viricel & Rosel, 2012). Additionally, the mitochondrial control region has been evaluated for estimating the population genetics of whales (Arnason et al., 1993; LeDuc et al., 2017). Combining various nuclear and mitochondrial loci has proved useful in identifying whale phylogeny and divergence periods (Jackson et al., 2009; McGowen et al., 2009; Sasaki et al., 2005). Whole-genome sequencing of blue whales and other rorquals has revealed evidence of introgressive gene flow in the marine environment, as well as insights on their lifespan and adaptability (Árnason et al., 2018; Yim et al., 2014). Therefore, this study aims to identify a morphologically unknown rorqual by analyzing mitochondrial DNA. To confirm the species’ identity, we calculate genetic distances and look at the phylogenetic relationship. Our generated data may serve as a reference sequence for population genetic analyses of rorquals in Indian marine waters. Additionally, historical records of blue whales spanning ~90 years will enhance our understanding and contribute to long-term conservation action plans for cetaceans in the Indian marine ecosystem.
Materials and Methods
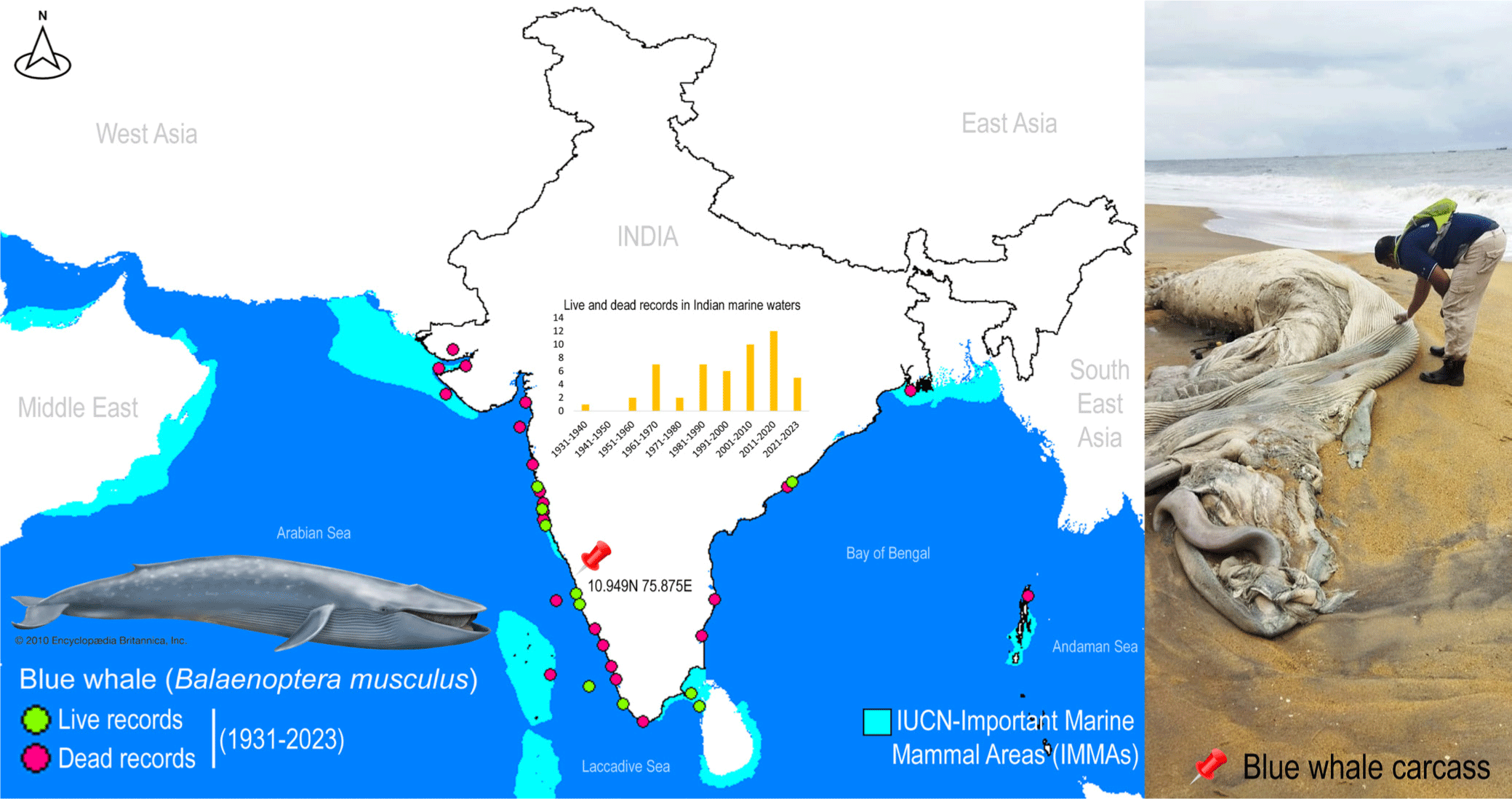
A single deceased individual of rorquals was detected along the western coast of India, near Kozhikode, Kerala, at coordinates 10.949N 75.875E (Fig. 1). The external morphological features of the specimen were extensively degraded, rendering taxonomic-based species identification unfeasible. To mitigate potential surface contamination, an aseptic tissue sample was carefully extracted and thoroughly rinsed with 70% ethanol before preservation. The tissue sample was vouchered and cataloged in the National Zoological Collections of the Western Ghat Regional Centre (WGRC), Zoological Survey of India (ZSI), Kozhikode, India, under registration number ZSI/WGRC/V.3628. In an effort to gain insights into the conservation priorities for rorquals in Indian marine waters, Important Marine Mammal Area (IMMA) shapefiles and associated metadata were acquired from the IUCN Joint SSC/WCPA Marine Mammal Protected Areas Task Force (IUCN MMPATF, 2018).
Total genomic DNA was extracted using the standard phenol–chloroform isoamyl alcohol method (Sambrook & Russell, 2001). Two primer pairs, LCO-HCO and mcb398-mcb869, were employed to amplify the partial segments of COI and Cytb genes, respectively (Folmer et al., 1994; Verma & Singh, 2002). Each 25 mL PCR reaction mixture contained 10 pmol of each primer, 20 ng of template DNA, 1X PCR buffer, 1.0–1.5 mM of MgCl2, 0.25 mM of each dNTPs, and 1 U of High-fidelity Platinum Taq DNA Polymerase (Invitrogen, Carlsbad, CA, USA). PCR reactions were carried out in a Veriti Thermal Cycler (Applied Bio Systems, Foster City, CA, USA) following standard thermal profiles. PCR products were purified using the QIAquick Gel Extraction Kit (QIAGEN, Germantown, MD, USA) according to the provided protocol. Cycle sequencing was performed with the BigDye Terminator ver. 3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA), and bi-directional Sanger sequencing was conducted using the Genetic Analyzer (Applied Biosystems) located at Rajiv Gandhi Centre for Biotechnology, Thiruvananthapuram, Kerala, India.
Forward and reverse chromatograms were carefully screened using the SeqScanner Version 1.0 (Applied Biosystems) to eliminate noisy segment from each sequence. Consensus sequences were subsequently subjected to nucleotide BLAST searches (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 21 July 2023) and ORF finder screening (http://www.ncbi.nlm.nih.gov/gorf/gorf.html, accessed on 21 July 2023) for validation. Finally, the generated sequences were deposited in the GenBank database to obtain unique accession numbers. Additionally, a total of 43 COI sequences and 35 Cytb sequences from various Balaenopteridae species were retrieved from GenBank to facilitate confirmatory analyses. The DNA sequence (NC_000889) of Hippopotamus amphibius (Artiodactyla: Hippopotamidae) was used as an outgroup taxon in the ensuing analyses. Generated and database sequences were aligned using ClustalX software to create two distinct datasets (Thompson et al., 1997).
Kimura-2-parameter (K2P) genetic distances were computed for both COI and Cytb datasets using MEGAX (Kumar et al., 2018). To conduct phylogenetic analyses, the best fit models were determined using JModelTest v2 based on the lowest Bayesian Information Criterion scores (Darriba et al., 2012). Bayesian inference topologies were constructed using Mr. Bayes 3.1.2, employing nst = 6 with one cold and three hot chains (Metropolis-coupled Markov chain Monte Carlos). The program was executed for 100,000 generations, with tree sampling performed at every 100 generations and 25% of samples discarded as burn-in (Ronquist & Huelsenbeck, 2003). The resulting topologies were edited using the iTOL v6.8 webserver (https://itol.embl.de/login.cgi, accessed on 21 July 2023) (Letunic & Bork, 2021).
Results and Discussion
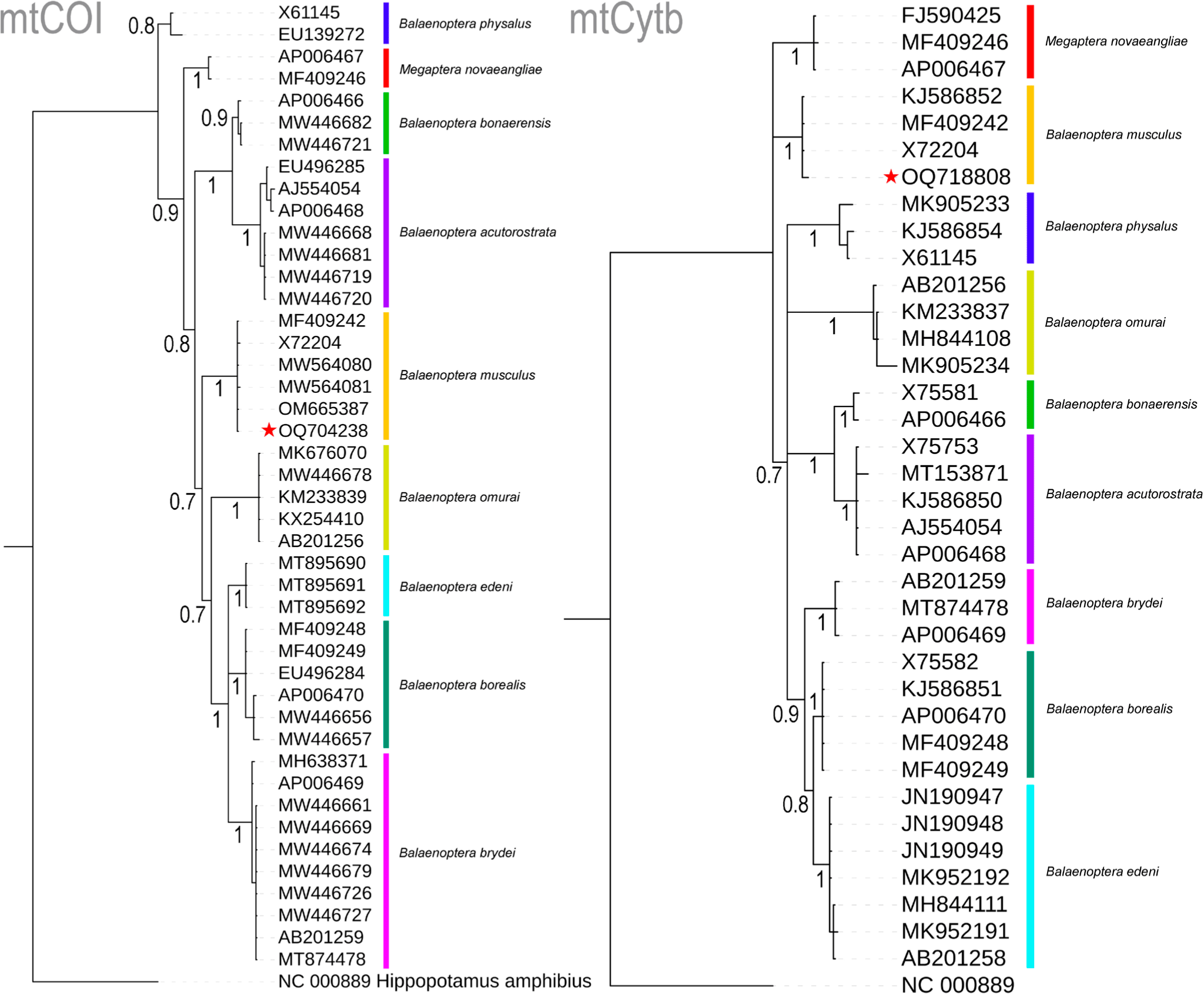
The four nucleotides (A, T, G, and C) wield significant power in identifying organisms across various life forms and stages, particularly when morphology alone proves inadequate for the task (Hebert et al., 2003). This innovative approach, seamlessly integrated with traditional morphology-based species identification, has demonstrated its effectiveness, particularly in the context of Marine Metazoans (Steinke et al., 2016). In the present study, molecular techniques were employed to examine a deceased rorqual specimen. The generated sequences of two mitochondrial genes, COI (OQ704238) and Cytb (OQ718808), exhibited striking similarities of 100% and 99.56%, respectively, with the references mitochondrial genome sequence (MF409242) of B. musculus (Árnason et al., 2018). Among the nine studied Balaenopteridae species, the partial COI gene (612 bp) displayed 20.75% variable nucleotides, with a mere 0.33% variability observed in B. musculus. Similarly, the partial Cytb gene (450 bp) exhibited 24.22% variable nucleotides, with 0.44% variation in B. musculus. Both COI and Cytb genes depicted mean genetic distances of 7% and 8%, respectively, within the present datasets. Specifically, in the COI gene, inter-species genetic distances ranged from 3.83% (between B. bonaerensis and B. acutorostrata) to 10.42% (between B. borealis and B. acutorostrata) (Table 1). Notably, the targeted species, B. musculus, including the generated sequence, maintained inter-species genetic distances of 6.75% (with B. borealis) to 9.80% (with B. acutorostrata) when compared to other Balaenopteridae species. Intra-species genetic distances among the Balaenopteridae species ranged from 0% (between B. edeni and B. omurai) to 1.2% (for B. physalus). In the Cytb gene, inter-species genetic distances spanned from 2.66% (between B. borealis and B. edeni) to 12.78% (between Megaptera novaeangliae and B. bonaerensis) (Table 1). The blue whale, B. musculus exhibited inter-species genetic distances ranging from 7.37% to 10.96% when compared to other Balaenopteridae species. In contrast to COI, the Cytb gene displayed negligible intra-species genetic distances, ranging from 0% to 1.1%, as observed in COI. Both COI- and Cytb-based Bayesian phylogenies exhibited distinct segregation and monophyletic clustering for all Balaenopteridae species. The generated sequences from the unidentified deceased rorqual formed cohesive clusters within the B. musculus clade in both COI- and Cytb-based species trees, with robust posterior probability support (Fig. 2). Based on this molecular investigation, the present study confirms the identity of the deceased rorquals as blue whales, B. musculus.
Despite their crucial role in marine ecosystems, whales, and especially in India, have received scant attention from conservation biologists and taxonomists (Bijukumar et al., 2012). Marine mammals frequently strand along the Indian coast, yet due to a dearth of local taxonomic expertise and the often-compromised condition of specimens, accurate species identification is not consistently achieved (George et al., 2011). Given the pivotal role of all cetaceans in conservation endeavors, precise documentation of their presence can yield vital insights into the distribution and migratory patterns of numerous species in the oceans surrounding India. The blue whale, B. musculus (cf. Linnaeus 1758), stands as the largest baleen whale and indeed, the largest known animal on Earth. This colossal mammal boasts a cosmopolitan distribution across major oceans worldwide, with the exception of the Arctic, Mediterranean, Okhotsk, and Bering Seas. It encompasses four recognized subspecies, namely B. m. musculus, B. m. indica, B. m. intermedia, and B. m. brevicauda (Reeves et al., 1998). In the Indian Ocean, blue whales (B. m. indica) have been recorded on numerous occasions along both the east and west coasts, from Okhamandal in Gujarat to Andhra Pradesh and West Bengal states in India. These records are based on strandings, acoustic detections, and live sightings (Chandrasekar et al., 2021; Marine Mammal Research & Conservation, 2023; Sutaria et al., 2016). It is worth noting that the blue whale (B. musculus) is currently categorized as ‘Endangered’ by the IUCN Red List of Threatened Species (IUCN, 2023).
Blue whales face a myriad of anthropogenic threats that hinder their population recovery, including historical whaling, climate-induced pollution, noise pollution, and vessel collisions. The causes of marine mammal strandings can be attributed to both natural and human-induced factors, and the information gleaned from strandings can be instrumental in understanding the geographical distribution of marine mammal populations, their seasonal migrations, and the factors contributing to mortality. While the effects of noise pollution on blue whales are not yet fully understood, it has the potential to disrupt mating activities, leading to population-level consequences and harm or mortality (Croll et al., 2001). Although a recent study has focused on a significant mass mortality event among large baleen whales, linked to harmful toxic algal blooms (Häussermann et al., 2017), the natural causes of mortality among blue whales are well-documented. However, the specific reasons for mortality in stranded marine animals are often inadequately studied, and extensive necropsy research and postmortem examinations would greatly contribute to understanding the impact of anthropogenic interactions. Blue whales, like other baleen whales, undertake extensive migrations between breeding areas in low-latitude regions where they mate and give birth during the winter, and feeding grounds in high-latitude areas during the summer (Corkeron & Connor, 1999). The energy expended during these activities, including fasting, migration, calving, and raising young, increases the risk of mortality during these voyages. The southern range of the Indian Ocean boasts the largest acoustic diversity of blue whales (Torterotot et al., 2020). Given the recent establishment of IMMAs in Indian marine waters, both the east and west coasts feature high-priority zones, representing critical habitat portions for marine mammal species that have the potential to be delineated and managed for conservation purposes. Beyond the realm of DNA data, blue whale sightings in Indian seas are on the rise, underscoring India’s ongoing conservation efforts. Furthermore, the Marine Mammal Research and Conservation Network of India (http://www.marinemmals.in/) has compiled the most comprehensive information on Indian cetaceans, undoubtedly facilitating conservation management planning.







