Introduction
In the Asia Pacific region, mud crab (Scylla spp.) has great economic value and becomes a potential candidate for cultured biota in brackish water ponds due to its high growth rate (Gunarto & Herlinah, 2015; Gunarto et al., 2020; Talib et al., 2017). Thus, it is necessary to produce mass mud crab seeds continuously to fulfill the needs of the crab seeds. However, crablet production from hatcheries is still low (Gunarto et al., 2021; Waiho et al., 2018) due to the mass mortality of Scylla spp. larvae frequently occur after several days of rearing. The mass mortality of larvae is due mainly to pathogenic bacterial infections. Vibrio harveyi, Vibrio alginolyticus, and Vibrio parahaemolyticus are pathogenic bacteria that have been reported to infect mud crab larvae (Talib et al., 2017). To promote better survival rates of cultured biota, including in larvae of Scylla spp. culture, utilization of beneficial bacteria, so-called probiotics, has been performed.
Probiotic bacteria have already been widely applied in aquaculture practices (Kuebutornye et al., 2019). Specific probiotic isolates can generate digestive enzymes that support healthy gut bacterial communities, inhibit harmful bacteria formation, and boost overall immunity. Water quality in hatcheries has reportedly been improved by introducing probiotics directly to the water during larval culture (Ayisi et al., 2017; Buruiană et al., 2014; Hlordzi et al., 2020). There are many Bacillus spp. species that have been used as probiotics. As probiotics, Bacillus subtilis and Bacillus pumilus isolated from crab intestines can control pathogenic Vibrios in mud crab aquaculture (Wu et al., 2014). Furthermore, Bacillus amyloliquefaciens was isolated from the digestive tract of freshwater fish Labeo calbasu (Kavitha et al., 2018). Bacillus licheniformis supplemented feed fed prawns to be competitively excluded pathogenic bacteria, such as Pseudomonas spp., Klebsiella spp., Staphylococcus spp., and Acinetobacter spp. in shrimp gut (García-Medel et al., 2020; Sudha et al., 2019). B. subtilis, a beneficial bacterium, has been applied in shrimp and fish aquaculture, improving water quality (Hlordzi et al., 2020; Olmos, 2014).
The ideal dose of B. subtilis as probiotics for the mud crab Scylla spp. larvae rearing in the hatchery, however, is not well understood. Beneficial probiotics application in larvae rearing is expected to reduce the risk of developing various diseases, including Vibriosis caused by Vibrio spp. infection and to optimize the larvae growth to the crablet stage. In the present study, we use probiotics containing B. subtilis, which was initially isolated from seaweed, Kapaphycus alvarezii, that was cultured in the coastal area of Laikang village, Jeneponto regency, South Sulawesi, Indonesia. Probiotics have also been kept in powder or powder form to make their application easier and shield probiotic organisms from environmental damage. The mechanisms of Bacillus spp. to eliminate pathogenic bacteria include one of the ways by the formation of macrophages extracellular traps as the central cell of immune response (Romo-Barrera et al., 2021). Earlier researchers reported that to prove the efficacy of probiotics-fed diets containing B. subtilis, the real-time PCR was employed to determine the mRNA levels of prophenoloxidase, peroxinectin, lipopolysaccharide- and b-1,3-glucan-binding protein and serine protein. The expression of all immune-related genes studied could be significantly up-regulated (p < 0.05) than control (Zokaeifar et al., 2012).
Probiotics can also alter the gut microbiota of larval, fry, and juvenile intestines to reduce pathogen adherence and colonization. To increase the quantity of good bacteria in the stomach, they are introduced to water or a fish diet (Vaseeharan & Ramasamy, 2003). Due to the small size of megalopa, we were not able to take their intestine for samples of gut microbiota; therefore, the whole body of megalopa and water media for larvae rearing are used for NGS analysis for observation on the effect of different dosages of probiotics B. subtilis to the microbiota composition and abundance. Our study aims to study the effect of various doses of probiotics on the mud crab Scylla tranquebarica larvae rearing. In addition, we also looked at the number of microbiotas in the megalopa’s entire body and the water used for larvae rearing, water quality parameters, larvae development, and crablet production. To generate the best crablets possible in the hatchery, we must also identify the suitable doses of probiotics containing B. subtilis to be applied during the rearing of S. tranquebarica larvae.
Materials and Methods
Seawater at a salinity of 30 ppt was sterilized using 20 mg/L chlorine overnight, then aerated and neutralized with 20 mg/L sodium thiosulfate the following night (Syafaat et al., 2019). Subsequently, the water was filtered using a filter membrane and through Ultraviolet light before being transferred to the 200 L volume of conical fiber tanks used for rearing the larvae. Before the larvae were stocked in the cone fiber tank, 15 mg/L of erythromycin was added, which is equal to three grams in each tank (Gunarto et al., 2021). Antibiotics were applied earlier before larvae stocked aimed to cut the vertical transmission of pathogenic bacteria from the broodstock to their larval and horizontal transmission from live food to the gut of larvae (usually Vibrio spp.), which mainly causes death in mud crab larvae. Furthermore, the tanks were sufficiently aerated to supply oxygen to the larvae. The S. tranquebarica larvae, seen swimming on the surface of the water when freshly hatched and in healthy condition, were put into cone fiber tanks at an abundance of 80 ind/L. From the first to the seventh day, rotifers, Brachionus spp. at the density of 20 ind/mL enriched with highly unsaturated fatty acids (HUFA) at concentrations of 100 mg/L (DHA Selco, INVE Aquaculture, Salt Lake City, UT, USA). Kotani (2017) was fed to the larvae. Moreover, starting on the seventh day of the rearing, the larvae were fed with Artemia nauplii with a density of 0.5 ind/mL enriched with 500 mg/L HUFA for six hours. Conversely, the larvae were exclusively fed Artemia nauplii at a density of 1 ind/mL on the tenth day. Finally, the number of Artemia nauplii increased to 1–2 ind/mL in the megalopa stage until they developed to the crablet stage. In rearing water, ammonia, nitrites, and total organic matter (TOM) were measured by collecting 300 mL of water from each replication tank. Those parameters were analyzed according to Clesceri et al. (2005). Meanwhile, YSI Pro Plus multiparameter water quality (RoHS CE WEEE C-Tick VCCI FCC IP-67 1-meter drop test Assembled in the USA) is used to measure dissolved oxygen (D.O.), pH, and water temperature.
The probiotic containing B. subtilis was produced by the Research Institute for Brackish Water and Fisheries Extension, Maros, where the B. subtilis isolated from seaweed, K. alvarezii, which grows in the coastal area in Laikang, Jeneponto regency, South Sulawesi, Indonesia. That probiotic is made in powder form with a B. subtilis density of 108 CFU/g. This probiotic was administered in the mud crab larvae rearing at the following doses A) 2.5 mg/L, B) 5.0 mg/L, C) 7.5 mg/L, and D) 0.0 mg/L (control without probiotic application) by giving the probiotic powder at 0.5, 1.0, 1.5, and 0 g, respectively per 200 L of rearing water. The probiotics need to be activated before being added to the larvae-rearing tank. The probiotic powder was placed in a small basin with a volume of three liters, then filled with two liters of sterile seawater at a salinity of 30 ppt and aerated overnight. On the next day, the aeration stopped. The solution was settled for one hour to let the particle sink. Then, only a clear solution is added to the mud crab larvae cultures. The probiotic administration began on the seventh day of larva rearing and continued with the interval of three days until the larvae reached the first instar crablet stage (about 24–28 days).
The water exchange is as much as 10% of the total water volume in larvae rearing tanks and is first conducted on 12 days of rearing; furthermore, water exchange is continuous every three days later in the same volume until larvae growth to the megalopa stage. Again, in the megalopa stage, the water exchange was increased to 15% of the total volume every three days until megalopa developed into the crablet stage. The probiotic is also added to maintain probiotics in larval and megalopa rearing tanks as the dosage was determined to treat every after-exchange water in the larval and megalopa rearing tank. The sampling time for profiling the microbiota was conducted when larvae reached the megalopa stage at DOC 21 (first replication) and at DOC 22 (second replication) with the reason that the megalopa stage is the end stage before larvae develop to the crablet stage.
16S rRNA amplicon sequencing was carried out to elucidate microbial abundances in megalopa and media for larvae rearing treated with probiotics at different doses. The samples for the analysis were prepared as follows: First, megalopa extraction was conducted by taking ten individuals of megalopa from each replication of probiotic treatment and control tank. Then, the megalopas from each treatment were put in each sterile microtube, crushed, and extracted using the cTAB dTAB DNA extraction kit (IQ2000, GeneReach Biotechnology, Taichung, Taiwan). Then, the pelleted DNA was dissolved in 200 mL T.E. buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.5). The sample extract megalopa from each treatment was determined as A, B, C, and D. Each sample had two replications.
Genomic bacterial DNA from seawater samples was prepared by filtering one liter of water samples from the larvae-rearing tank in each treatment. The water was filtered using plankton net no. 400 with mesh size 37 µm to eliminate rotifer from the rearing water. The rotifer-free water was then filtered using Whatman filter paper Purabind 045, mesh size 0.45 micrometer, and diameter 47 mm. The filter paper was extracted following the ZymoBiomics DNA miniprep kit extraction procedure. The sample of extract water media for megalopa rearing from each treatment was determined as E, F, G, and H. Each sample had two replications.
Genomic DNA from megalopa and water samples were transported to the Genetics Science Laboratory (Jakarta, Indonesia) for the 16S rRNA amplicon sequencing in a PE250 Illumina sequencing platform. A DNA template with a concentration of 1 ng/μL was obtained by diluting the sample with sterile water after monitoring its concentration and purity on 1% agarose gels and measuring it using Nanodrop. The bacterial 16S rRNA gene’s V3-V4 region was amplified using primers 341F (5′-CCTAYGGGRBGCASCAG-3′) and 806R (5′-GGACTACNNGGGTATCTAAT-3′; Hjelmsø et al., 2014) with the barcode. The Phusion® High-Fidelity PCR Master Mix (New England Biolabs, Ipswich, MA, USA) was used for all PCR reactions, with a volume of 20 μL per reaction, under the following conditions: 0.4 μL TaqHiFi polymerase, 1 μL template cDNA, 4.0 μL 5 × buffer, 0.8 μL forward and reverse primer (5 μM), 2 μL dNTPs (2.5 mM), and 11 μL sterile water. The following were the amplification conditions: there were 25 cycles following pre-denaturation for two minutes at 95°C: 30 seconds of denaturation at 95°C, 30 seconds of annealing at 55°C, 30 seconds of extension at 72°C, and 5 minutes of final extension at 72°C (Sun et al., 2015). Equivalent densities of the PCR products were combined. Before sequencing the 16S rRNA amplicon, the mixed PCR products were purified using the Qiagen Gel Extraction Kit (Qiagen, Hilden, Germany). Then, the library was prepared using the NEBNext® UltraTM DNA Library Prep Kit for Illumina, and the quantification was done using Qubit and Q-PCR.
Samples were assigned paired-end readings based on their distinct barcodes, which were then shortened by removing the primer sequences and barcode. To get raw tags, FLASH version V1.2.7 was used to integrate paired-end reads. To get raw tags, FLASH version V1.2.7 was used to integrate paired-end reads (Magoč & Salzberg, 2011). The raw tags were filtered using QIIME (V1.7.0) to produce clean, high-quality tags (Caporaso et al., 2010). Using the UCHIME technique, the tags were compared with the reference database (gold database; Edgar et al., 2011) to identify sequence chimeras. Next, the chimeric sequences were eliminated.to detect chimera sequences (Haas et al., 2011) to get effective tags. Using Uparse version v7.0.1001, comparable operational taxonomic units (OTUs) were assigned to sequences with ≥ 97 percent similarity (Edgar, 2013). For each representative sequence, Mothur software was performed against the SSUrRNA database of SILVA Database (SILVA version 128, http://www.arb-silva.de/; Wang et al., 2007) for the annotation of species at each taxonomic rank (species, genus, order, family, kingdom, phylum, class, threshold: 0.8–1; Quast et al., 2013). The MUSCLE version 3.8.31 was used to determine the phylogenetic relationships of all OTU representative sequences (Edgar, 2004) and was employed to quickly compare several sequences. The OTU abundance data was standardized by assigning a standard sequence number to the sample with the fewest sequences. Based on the output normalized data, further analyses of beta and alpha diversity were carried out. Using indices like Chao1, Shannon, and Simpson, Alpha diversity was used to measure the species diversity complexity for a sample. With QIIME (version 1.7.0), all these indices in our samples were computed, and R software (version 2.15.3) was used to visualize the results including clustering of dissimilarity values of bacterial relative abundance using the complete linkage clustering method. Furthermore, the changes in samples’ species complexity, as well as those of individual samples and treatments, were assessed using the beta diversity analysis. QIIME was used to determine the beta diversity on weighted and unweighted Unifrac (version 1.7.0).
Bacterial analysis was conducted by observing total bacteria on tryptic soy agar (TSA) media, while the total Vibrio test utilized thiosulfate citrate bile salt sucrose agar (TCBSA) media (1). From each replication tank, a 50 mL water sample was taken in a sterile dark bottle for analysis. Subsequently, a 1 mL water sample was mixed with 9 mL saline solution (0.85% (w/v) NaCl) and serially diluted four times (10–1; 10–2; 10–3; 10–4). For the TCBSA plate medium, 100 µl of the water sample without dilution and with the first dilution (10–2) was spread. The same procedure was followed for sample inoculation in TSA media using the third and fourth dilutions (10–3 and 10–4). The samples were evenly spread across the surface with a sterile glass spatula and then incubated for 48 hours. Manual calculations were performed on bacterial colonies grown in TCBSA (total Vibrio) and TSA media (total bacteria) using the formula:
P = bacterial population (CFU/mL), Q = total number of bacteria in one dilution level (colony), T = number of plates used, S = dilution rate, V = plate sample volume (0.1 mL; Buller, 2004).
Larvae samples were taken on three distinct surfaces in each replication tank using a 100 mL bowl to monitor the larvae population. The larval population density per liter was calculated from the mean larval number per 100 mL. In each replication tank, the megalopa population was observed using a 1,000 mL bowl placed on three different surfaces. The mean megalopa number per 1,000 mL indicated the megalopa population per liter. The number of crablets produced per replication tank in each treatment was also monitored after the crab was harvested.
Data on the relative abundance of microbiota from the amplicon sequencing in each treatment were made graphically and analyzed descriptively. We performed alpha and beta diversity as well as hierarchical clustering to understand bacterial community composition (BCC) among samples. The larval population and crablet production data in each treatment were subjected to logarithmic transformation. Furthermore, analysis of variance was used to compare and examine the data for the fully randomized design patterns. If there was a difference between the treatments, the least significant difference test (LSD test) was used. The statistical test was conducted using the SPSS (Statistical Product Service Solution) program package 23. In comparison, the data on water quality (ammonia, nitrite, TOM) from each treatment were analyzed descriptively.
Results
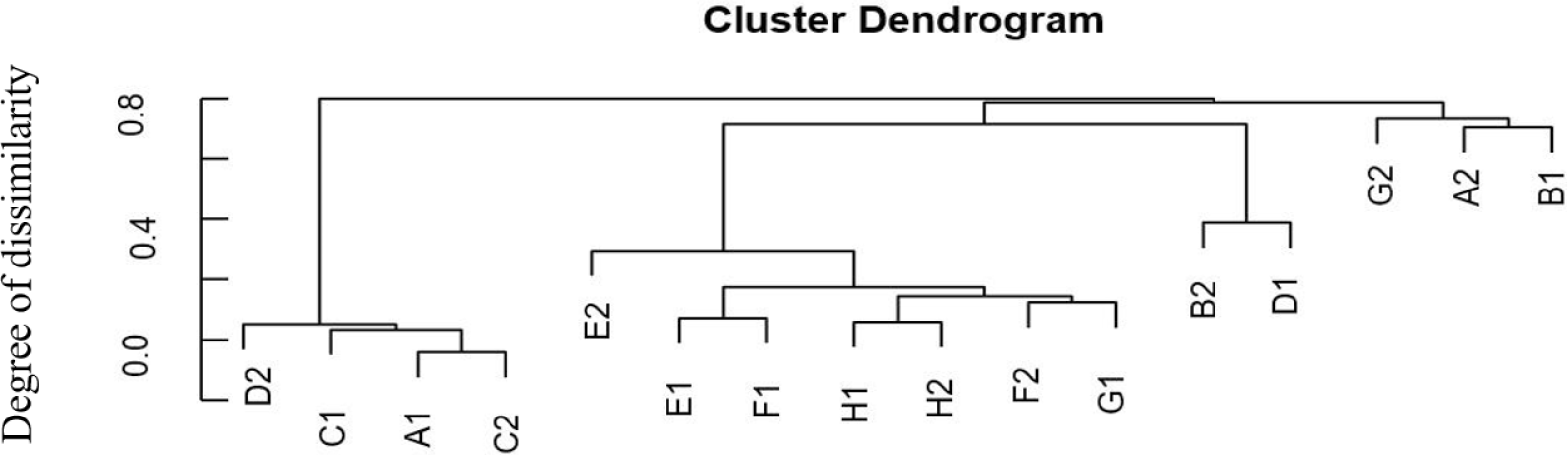
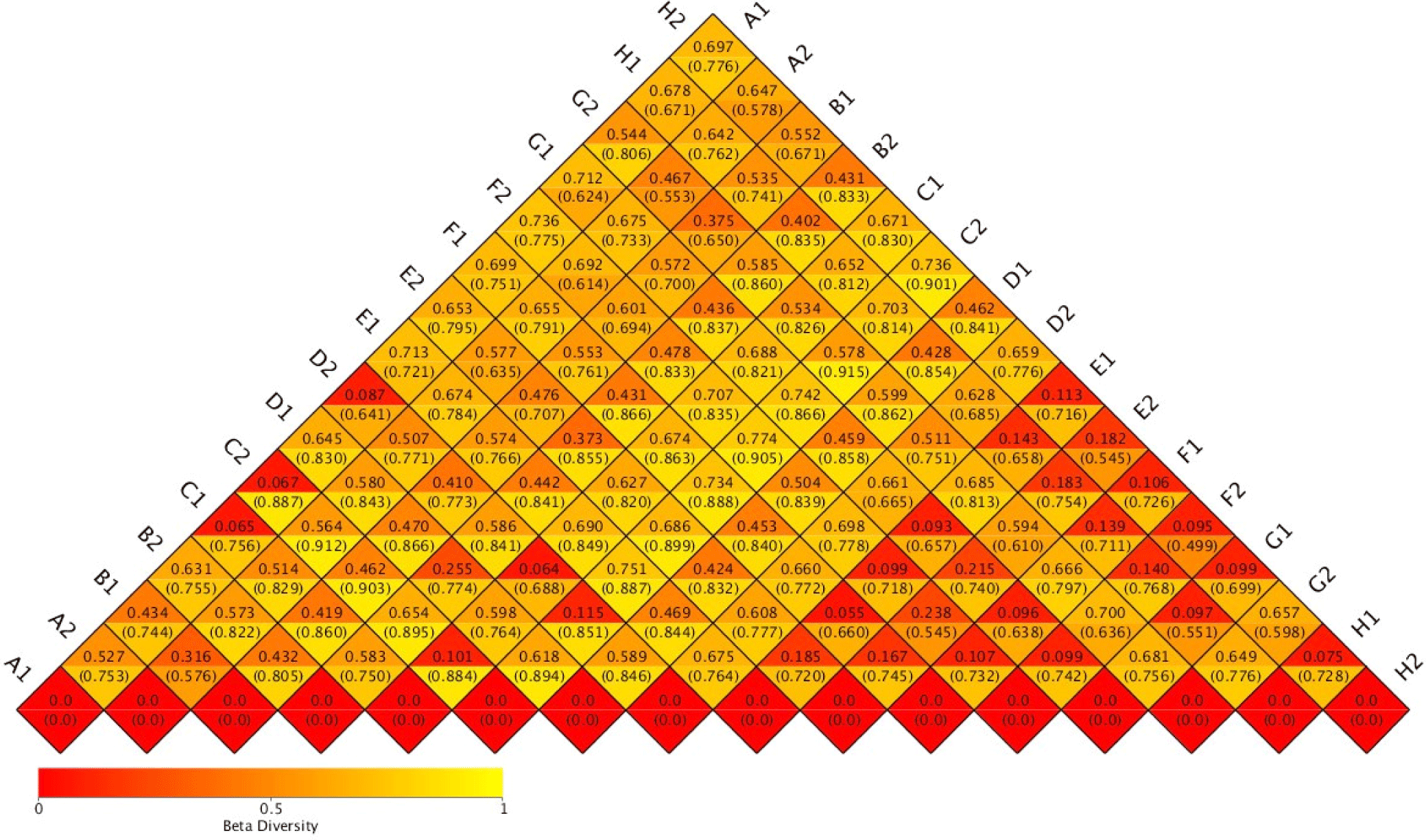
Species richness varied in the range of 269 to 1,052 OTUs. Despite relatively high species richness, each replicate within the treatment had twice a different number of species richness (Table 1). A hierarchical clustering for each sample based on the dissimilarity values of bacterial relative abundance showed four clusters (Fig. 1). BCC in water samples from all treatments clustered together. They were also separated from the BCC of megalopa samples at any dose of probiotics. Furthermore, different clustering patterns within similar treatments of replicate samples, except treatment C samples indicated that the replicate samples had relatively high dissimilarity value (> 0.7; Fig. 2).
The heatmap displaying the distribution of the top 35 microbiota genera over megalopa’s entire body and the water media for megalopa rearing after being treated with different dosages of probiotics containing B. subtilis is shown in Fig. 3 . The composition of microbiota in megalopa and the water media at the highest abundance is defined as (+3) and at the lowest abundance is described as (–3). The data showed that the highest abundance of microbiota in the whole body of Megalopa with a value between +2 and +3 includes Raistonia found in samples of A1, C1, C2, and D2. Pseudomonas in a sample of A2, Megaspaera found in a sample of B1, and Enterobacter, Corynebacterium, Cutibacterium, Streptococcus, Enhydrobacter, Micrococcus found in a sample of B2. Oceanospirillum, Marinibacter, Halomonas, Rothia, Methylolligelaceae, Photobacterium found in sample of D1. While the lowest abundance of microbiota in the whole body of Megalopa with the value –1 to –2 found in samples of A1, A2, B1, C1, C2, and D2 includes the genera of Ruegeria, Saprospiraceae, OM43_clade, Phaeodactilobacter, and Salinihabitants.
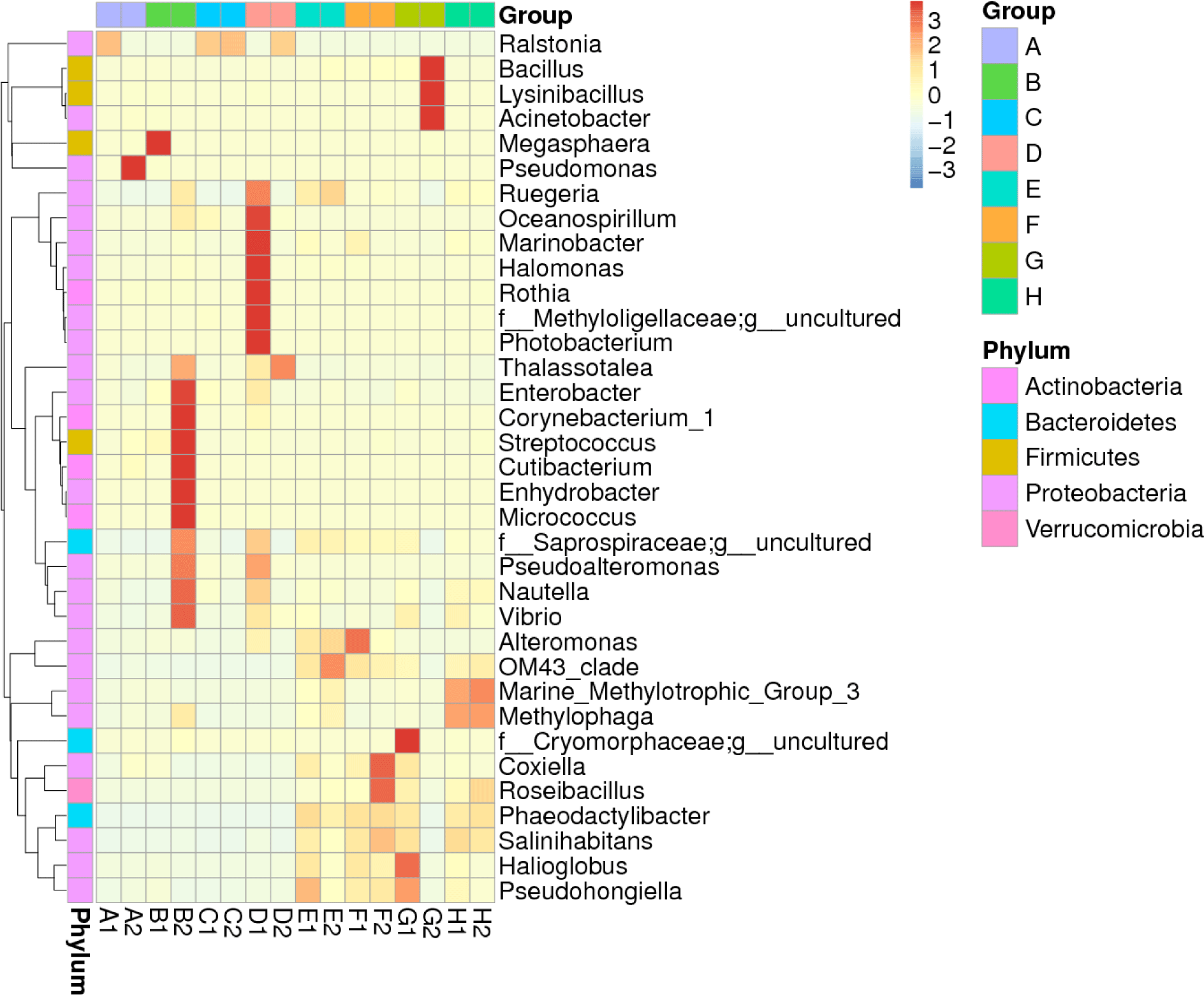
The highest abundance of microbiota in the water media with values of +2 to +3 includes the genera of Bacillus, Lycinibacillus, and Acinetobacter found in samples of G2. While Pseudohongiella in a sample of E1, OM43_clade in a sample of E2, Alteromonas, OM43_clade in a sample of F1, Coxiella, Roseibacillus, Phaeodactylibacter, Salinihabitans, Pseudohongiella, and Halioglobus in the sample of F1, F2, G1. The Vibrio showed low abundance with the value –1 to –2 found in samples of A1, A2, B1, C1, C2, E2, F2, and G2.
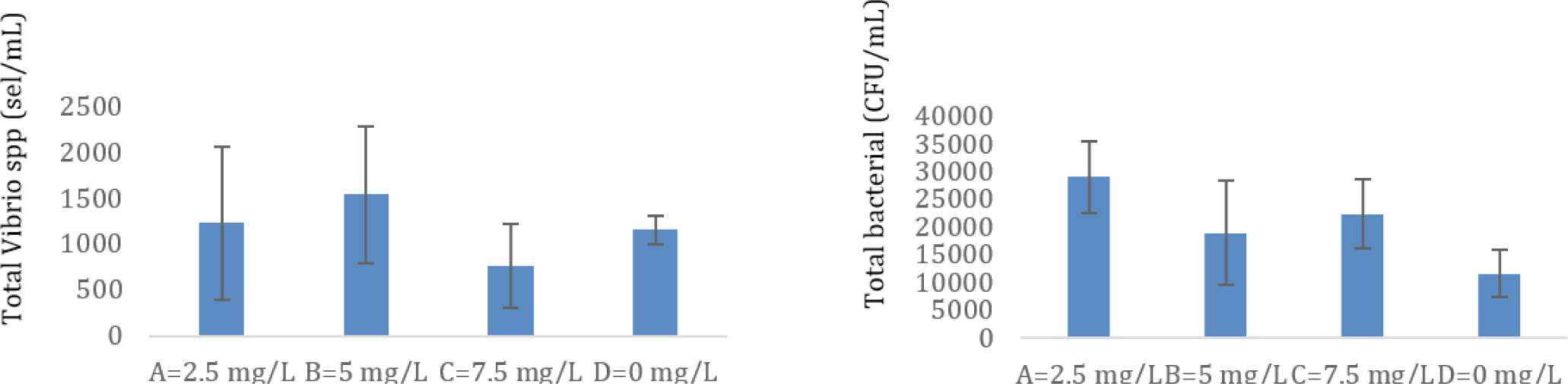
The population of Vibrio sp. and the total bacteria in the megalopa stage are shown in Fig. 4. It appears that the lowest Vibrio sp. population was found in treatment C (7.5 mg/L), although the total bacteria in treatment C was not the highest. The highest total bacteria were in treatment A (2.5 mg/L), and the lowest total Vibrio spp. did not occur in treatment A. The lowest total bacteria are in treatment D. The ratio of total Vibrio spp. to total bacteria was higher in control (0.1), compared to the treatment tested, namely of 0.035 (treatment C), 0.088235 (treatment B), and 0.033333 (treatment A).
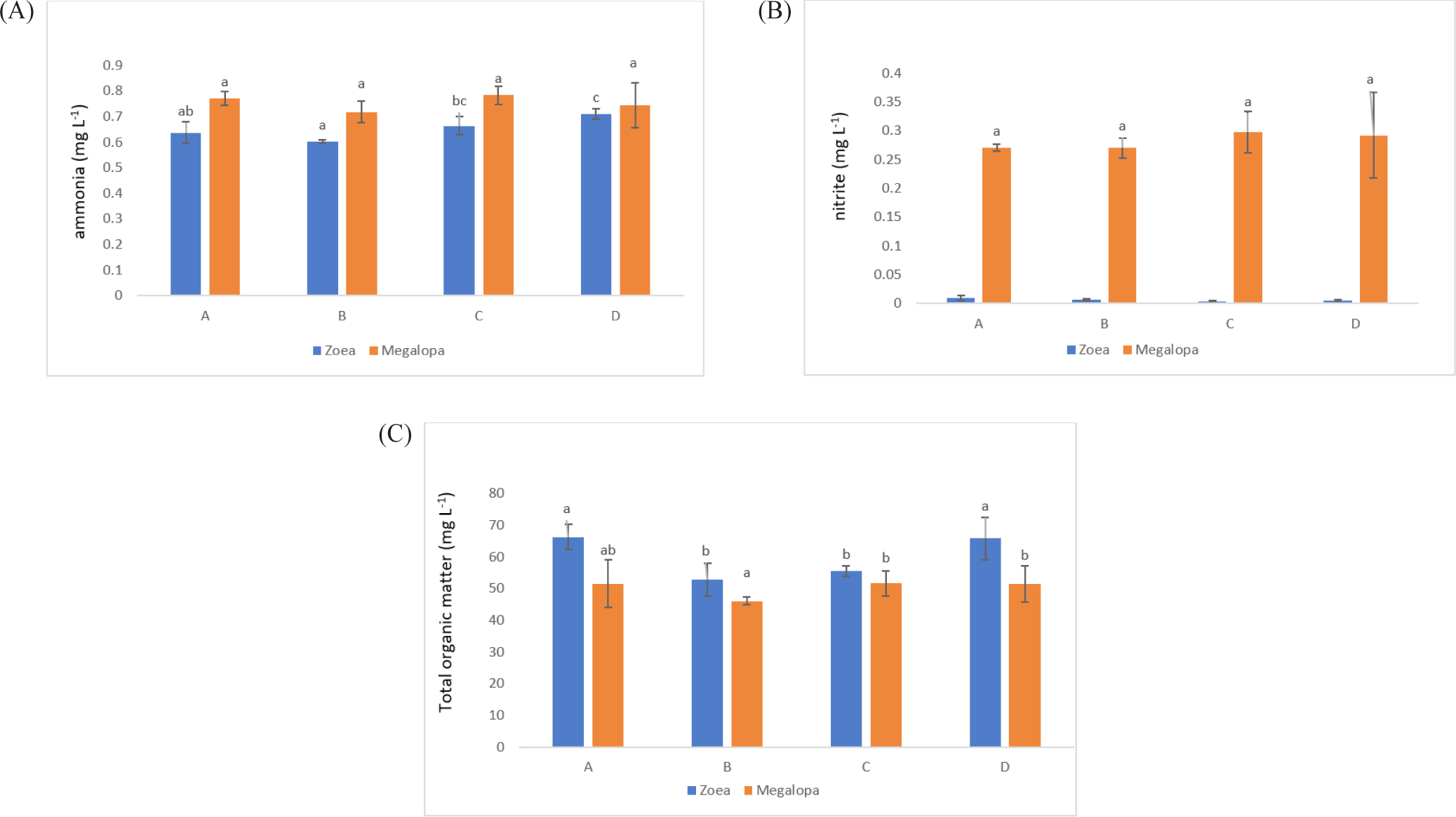
The salinity for larval rearing was 30 ppt, D.O. in the range of 5.2 mg/L to 6.7 mg/L, water pH: 8.1 to 8.4, and water temperature in the tank 27.5°C to 30.2°C. The ammonia concentration decreased at 10.3% (A), 15.23% (B), and 6.48% (C) compared to control (D) in the zoea stage. In contrast, the ammonia concentration significantly reduced to 3.45% (B) at the megalopa stage compared with the control treatment (Fig. 5A). All treatments’ nitrite concentration at the zoea stage was relatively low, reaching 0.01–0.07 mg/L. At the megalopa stage, all treatments’ nitrite concentration was increased (Fig. 5B). The TOM in the zoea stage decreased to 23.63% and 19.40% in treatments B and C, while at the megalopa stage, the TOM decreased to 4.34% only in treatment B, compared with the D treatment (control) (Fig. 5C).
The larvae population declined from zoea-1 to zoea-5. On the seventh and seventeenth days, the larvae in treatment C were significantly larger populations (p < 0.05) than those in treatments A and D, while on Day 17, a considerably larger population (p < 0.05) for the larvae in treatment B compared to the larvae in the control treatment. On day 23, treatments B and C had more megalops than treatments D. Treatments B and C yielded significantly (p < 0.05) more crablets than treatments A and D after crablet harvested from the tanks at the Crablet day 17 (Table 2).
Discussion
The application of different doses of probiotics, containing B. subtilis at the doses of 2.5 mg/L, 5 mg/L, 7.5 mg/L, and 0 mg/L in this research did not dramatically affect BCC in water samples. It was indicated by a low ß-diversity of the bacterial community in the water samples (Table 1). We supposed that the viability and adaptability of the probiotics B. subtilis was low as it was shown only one sample (G2) which contained the highest dose of B. subtilis. Our finding is similar to Amin et al. (2023). The microbiota, such as Ralstonia, highly increases population mostly at the megalopas reared with the addition of probiotics at 2.5 mg/L and 7.5 mg/L and control (Figs. 3 and 4). It was grouped in the first cluster (Fig. 1), where Ralstonia spp. is 90% dominant in these megalopas, and it was impacted to the lowest of Vibrio spp. population in megalopa. Ralstonia is a bacterium that can survive in highly purified water and disinfectant. It can be a used carbon source; therefore, bacteria can be a candidate for bioremediation (Adley et al., 2005). This activity may also cause improved water quality in larvae-rearing tanks. Besides that, Bacillus is also highly abundant in water media for megalopa rearing (sample G2) and the lowest of the Vibrio spp. population. All of them were impacted to the highest larvae survival in treatment C and ended with higher crablet production in treatment C.
In treatments B and C, applying probiotics at dosages of 5 mg/L and 7.5 mg/L means higher levels of beneficial bacteria were added to the larvae-rearing tank in those treatments. Reducing the growth and proliferation of pathogenic bacteria in the stomach larvae can be achieved by consuming high amounts of specific types of helpful bacteria (Buruiană et al., 2014), such as B. subtilis in the present study. That means megalopa in treatments B and C consumed a higher number of B. subtilis compared to the megalopa in treatments A and D, since the beginning of probiotic application have seen that the zoea population was higher in treatment B and C compared to treatment A and D and finally resulted in higher crablet production in treatment B and C (Table 2). Acinetobacter and Lysinibacillus also increased significantly in the water after being treated with probiotics at 7.5 mg/L. Phaeodactylibacter, Salinihabitans, Halioglobus, and Pseudohongiella distributed in all water media for megalopa rearing are applied with different dosages of probiotics (Fig. 3).
The diversity of microbiota populations in crab intestines is dominated by Bacteroides, Tenericutes, Cyanobacteria, and unknown bacteria (Li et al., 2016). Also, it was reported Firmicutes are absent in the intestines of sick crabs. However, Cyanobacteria is present. Meanwhile, in healthy crabs, Cyanobacteria are absent. In crabs from nature, the total content of bacteria in their intestines is more than 3–4 times the amount in sick crabs. In addition, the microbiota associated with crabs, for example, those in their intestines such as B. subtillis and B. pumilus are potential probiotics and can increase the immunity levels of crabs (Sahandi et al., 2019; Wu et al., 2014). Furthermore, the dominance of microbiota in shrimp, crab, and oyster cultured in ponds in Ganyu, Eastern, China dominated by Phylum Proteobacteria, Chloroflexi, Actinobacteria, Firmicutes, and Acidobacteria where water comes from the sea (Li et al., 2016). While in the intensive and semi-intensive pond in Indonesia, the halophilic bacteria (Alteromonas, Erythrobacter, Halomonas, Pseudoalteromonas, Salegentibacter, Sulfitobacter, and Psychrobacter) is dominant (Alfiansah et al., 2018). In the present study, the water used for larvae rearing was also from the coastal waters in Barru Regency, South Sulawesi, Indonesia. Based on the sequencing analysis, the microbiota in the whole body of megalopa and the water media for megalopa rearing was dominated by Proteobacteria, Actinobacteria, and small portions of Firmicutes, Bacteroidetes, and Verrucomicrobia. Gammaproteobacteria is the primary class in the Phyla of Proteobacteria with members including Aeromonadaceae, Pseudomonadaceae, Enterobacteriaceae, and Xanthomonadaceae (Li et al., 2016). B. subtilis belongs to phylum Firmicutes, thus in the water media for megalopa rearing, the population of Bacillus spp. was highest due to the application of probiotics at the highest dose (7.5 mg/L).
The efficacy of the bacteria, not their concentration, determines the potency of probiotic bacteria against pathogenic bacteria (Sahandi et al., 2019). In contrast to the present study, dosages of 5 mg/L to 7.5 mg/L resulted in higher crablet production than dosages of 2.5 and 0 mg/L. The higher concentration of probiotic bacteria applied in the larvae-rearing tanks affected the higher crablet production. In Lactobacillus sakei, the difference in secreted organic acid concentration determines the possible inhibitory level. The antibacterial activity of B. subtilis is mainly influenced by its ability to produce antibiotics, primarily peptides (Olmos, 2014; Stein, 2005).
In both human and veterinary medicine, erythromycin is a commonly used antibiotic that is effective against a broad range of bacteria. Erythromycin is also used in intensive livestock and aquaculture operations to promote growth and feed efficiency (Machado & Soares, 2019). The application of erythromycin in the present study aims to cut vertical transmission of pathogenic bacteria from the mud crab broodstock to their larvae. From our experiences, even though they have used erythromycin on the first rearing day, mass larvae mortality often occurs on days 10–12 caused by bacterial diseases. Therefore, applying probiotics, B. subtilis, from the seventh day of rearing could be initiated with the interval probiotic application every three days, which could finally increase the population of B subtilis in the larvae-rearing tank. Hopefully, it can prevent the development of pathogenic bacteria that often cause the larvae to die at day up to 10 days of rearing. Adding probiotic bacteria to the larvae-rearing medium can suppress the growth of Vibrio anguillarum in water media for larvae-rearing (Nogami & Maeda, 1992).
Additionally, the Bacillus probiotics can prevent the proliferation of pathogenic bacteria in the larval intestine (Buruiană et al., 2014), increase the microbiota population, and influence digestion, feed absorption, and assimilation (Ayisi et al., 2017). Unfortunately, the B. subtilis was unseen in the whole body of megalopa and water media by NGS sequencing analysis in the present study. However, the heatmap of the abundance of the top 35 genera of microbiota in the whole body of megalopa and in the water media for megalopa rearing after being treated with various doses of probiotics only showed Bacillus spp., which is abundant in water media, G2 at the dose’s probiotics application of 7.5 mg/L (Fig. 3). Whereas in other doses of probiotics treatments, the Bacillus spp. is lower density detected. The addition of antibiotics such as erythromycin may affect the microbiota composition and density in the whole body of megalopa and the water media of megalopa rearing in the present research. It was proven that the Bacillus spp. only seen a higher population in the water media of G2. However, the highest concentration (7.5 mg/L) of probiotic application was applied in the treatment of G and C (samples of G1, G2, C1, and C2). Therefore, those samples could have a higher density of Bacillus spp. Contrary, on G1, C1, and C2 samples unseen of Bacillus spp. presence. Furthermore, the most improvement in water quality especially for the ammonia and TOM was found in treatments B and C as a result of probiotics working in those treatments, and it was confirmed that treatments B and C have higher crablet production compared to those treatments A and D. Addition, an application of antibiotics affects reduce to the gut microbial diversity and composition (Patangia et al., 2022). B. subtilis will experience competitive stress when antibiotics like erythromycin are present, but they have systems to survive germs that develop resistance (Liu et al., 2018).
V. harveyi, is pathogenic to marine and inland fisheries biota (Alfiansah et al., 2018; Sahandi et al., 2019; Yilmaz et al., 2022), including larvae and adult mud crab (Aftabuddin et al., 2013; Poornima et al., 2012; Talib et al., 2017). Earlier researchers reported that V. harveyi attacks crab larvae by infecting the hemolymph and hepatopancreas (Poornima et al., 2012). Due to the open circulatory system in the crab, all parts of the body of the crab might have been attached to pathogenic bacteria (Zhang et al., 2020). Therefore, in this study, all parts of the megalopa (the whole body of megalopa) were used to observe the microbiota, including Vibrio spp. population by crushing to make raw DNA for sequencing analysis. Furthermore, the accumulation of organic material in the larvae-rearing tanks will trigger the increasing V. harveyi population in the water. Therefore, the raw DNA from the water for megalopa rearing was applied with different probiotic doses to observe the microbiota, including Vibrio spp. population in the present research. In addition, the Vibrio spp. in the present study detected with the NGS analysis showed a high (value +2), it was seen in samples of B2, while in samples of D1, G1, H1, and H2, the value was lower, approximately only +1 (Fig. 3). The other sample unseen the availability of Vibrio spp. That means treatments A and C are free from Vibrio spp. However, B2 is dense with Vibrio spp, but the larvae successfully developed into the crablet stage. It means an outbreak of Vibrio spp, which mainly causes larvae mortality in rearing tanks, is not incurred. Bacteria such as V. harveyi can change from non-virulent to virulent when they reach a certain density level or a dramatic environmental change (Zhou et al., 2012). Earlier researchers reported that B. subtilis could improve disease resistance, minimizing larval mortality caused by V. harveyi infection (Zokaeifar et al., 2012). Thus, applying probiotics containing B. subtilis bacteria is essential to eliminate V. harveyi infection and support the successful rearing of larvae to develop into the crablet stage.
Probiotics’ effectiveness or efficacy in larvae could be due to probiotics’ high intestine colonization capability, favorable development conditions, or larval resistance to foreign probiotics (Hlordzi et al., 2020). Therefore, B. subtilis supplementation in the current study may impact mud crab larvae’s improved immune response and disease resistance. It was confirmed with earlier research that supplementation with B. subtilis will enhance growth performance, immune response, and disease resistance to pathogenic bacterial infections (Buruiană et al., 2014). Probiotic bacteria have been proven to promote digestive function by generating digestive enzymes, resulting in better digestion and absorption of food and, as a result, increased feed utilization efficiency (Ghosh et al., 2016). B. subtilis is found in the intestines of crabs and can be used as a probiotic. A density of 105 CFU/g will increase immunity against V. parahaemolyticus infection in mangrove crab larvae and Scylla paramamosain (Wu et al., 2014).
The present research applied the probiotic containing B. subtilis at doses of 2.5 mg/L to 7.5 mg/L. Based on the total count of bacteria soluble in the water by TSA media analysis, the density of total bacteria in treatments B and C were found to be 1.85 × 105 CFU/mL to 2.2 × 105 CFU/mL compared to 1.6 × 104 CFU/mL in the control tank. Furthermore, the ratio of total Vibrio spp. to total bacteria was lower in the treatment test compared to the control test, which means total bacteria including probiotics bacteria applied in the larvae rearing can suppress the Vibrio spp. population in the larvae-rearing tank. The ratio of total Vibrio spp. to total bacteria in the shrimp pond during culture can be used as an early warning system to indicate the pond is in a dangerous condition when the ratio value in the treatment pond is up more than 10% of the control value (Susianingsih et al., 2022). In the present study, the ratio of total Vibrio spp. to total bacteria in treatment tanks, namely 0.035 (treatment C), 0.088235 (treatment B), and 0.033333 (treatment A), all of them are far smaller compared to the control (0.1). The lowest Vibrio spp. population was found in treatment C (7.5 mg/L), although the total bacteria in treatment C was not the highest population. It shows that probiotics in treatment C effectively suppress the population of Vibrio spp. While the highest total bacteria were found in treatment A (2.5 mg/L), unfortunately, the lowest total Vibrio spp. did not occur in treatment A. This means that the work of probiotics in treatment A is not as effective as the work of probiotics in treatment C. Other bacteria associated with water and crab larvae in treatment A may be responsible for it. The lowest total bacteria are in treatment D, it is appropriate because in treatment D there is no addition of probiotic bacteria (Fig. 4). Therefore, concentrations of probiotics at 5.0 mg/L to 7.5 mg/L are most effective in preventing the growth of the population of Vibrio spp. pathogenic in the larvae-rearing tanks. Treatments B and C produced higher populations of crablet (p < 0.05), in the amount of 225 ± 2.82 individuals/tank and 220 ± 16.97 individuals/tank, respectively. It was higher crablet production compared to the previous study, which was only 141.2 ± 34.1 individuals/tank using the application of erythromycin in their larvae rearing (Gunarto et al., 2021). The probiotic containing B. subtilis, presumably at a dose of 5.0 mg/L and 7.5 mg/L, produced the highest level capable of fighting harmful bacteria in mud crab larvae rearing, improving digestion in stomach larvae, and improving water quality for larvae rearing. Therefore, higher percentages of larvae successfully develop into the crablet stage.
During the zoea stage, the ammonia concentration in all probiotic treatments was lower concentration between 0.6 and 0.7 mg/L when compared to the control treatment, with the B treatment showing the lowest significance (p < 0.05). The ammonia concentration in all treatments was higher in the range of 0.7–0.8 mg/L during the megalopa stage due to the accumulation of organic material and other materials in the tank bottom. As larvae reach the megalopa stage, in other instances, the probiotics containing B. subtilis could have also accumulated more due to regular application in every three days. Regretfully, in the megalopa stage, the ammonia concentrations across the treatments did not differ significantly (p > 0.05). Because of excessive feed, the metabolism of the larvae, and the death of the rotifer and larvae in the rearing tank, B. subtilis works to metabolite ammonia produced from an accumulation of organic material. The first source of nitrogen from animal deaths or waste products is organic matter. Nitrosomonas and Nitrobacter, which are primarily involved in nitrification and occasionally denitrification, are responsible for nitrogen fixation (Liu et al., 2020). The process known as ammonification involves fungi or bacteria, such as Bacillus species, converting this organic nitrogen to ammonium (NH+4) and ammonia (NH3). The process known as nitrification involves the conversion of ammonium to nitrites (NO−2) and subsequently to nitrates (NO−3), primarily by Nitrosomonas and Nitrobacter species, respectively. Subsequently, nitrate is converted to nitrogen gas (N2) through a process known as denitrification, which eliminates bioavailable nitrogen and releases it back into the atmosphere (Bernhard, 2010). Through ammonification, nitrification, and denitrification, Bacillus species have contributed significantly to the nitrogen cycle. Certain bacteria, such as B. subtilis, use various forms of nitrogen, such as total ammonia nitrogen , N-NO3, N-NO2, and total Kjeldahl nitrogen, for their metabolism (Martínez-Córdova et al., 2015). This helps remove nitrogen from the water column. As a result, in comparison to a control treatment, the ammonia concentration at the zoea stage in the present study was lowered by 6%–15% (Fig. 5A). It is observed that the probiotic treatment has a lower nitrite concentration than the controlled treatment. They did not, however, differ significantly (p > 0.05; Fig. 5B).
The different forms of nitrogen in aquaculture wastewater can be eliminated by Bacillus species. Particulate matter and dissolved organic carbon accumulation are decreased by gram-positive bacteria. Both organic and inorganic carbon sources are used by heterotrophic bacteria as growth substrates. Because of this, they play a significant part in both the breakdown of organic matter and the creation of particulate food materials from dissolved organics (Padmavathi et al., 2012). Bacillus efficiently transforms organic matter into CO2, which is then used as a carbon source by β- and γ-proteobacteria. In contrast, other bacteria convert most of the organic matter into slime or bacterial biomass (Mohapatra et al., 2013; Zorriehzahra et al., 2016). As a result of recycling nutrients in the water column and lowering sludge accumulation, Bacillus are primarily used in aquaculture to remove the organic matter load (Soltani et al., 2019). Unused food is frequently linked to organic matter loads in aquaculture. However, probiotic Bacillus stimulates appetite by raising fish’s activity levels of digestive enzymes, which improves feed utilization and reduces waste generation (Hlordzi et al., 2020).
In the present research in comparison to the D treatment, the TOM concentration in the zoea stage was significantly lower (p < 0.05) in the B and C probiotic treatments. Additionally, it was lower by 19%–23.6% when compared to the control treatment. B treatment had a significantly lower TOM concentration in the megalopa stage (Fig. 5C). Finally, the probiotics containing B. subtilis added during larval rearing have been shown to improve water quality (ammonia, nitrite, and TOM) and have an impact on higher crablet production in treatments B and C. It was conformed with the Kuebutornye et al. (2019) and Olmos (2014) that probiotics containing B. subtilis added in larval rearing could improve water quality.
The LC-50 of unionized ammonia for zoea-1 and zoea-5 was at 4.05 mg/L to 6.64 mg/L after 24 hours (Neil et al., 2005). The nitrite in the zoea stage was in the range of 0.004 mg/L to 0.009 mg/L and increased in the megalopa stage, with the highest concentration at 0.21 mg/L to 0.35 mg/L in treatment D (control). The safe limits of nitrite in the larvae of Z1 to Z5 were in the range of 2.5 mg/L to 6.9 mg/L (Seneriches-Abiera et al., 2007). Thus, ammonia and nitrite concentrations in the present study still support larvae development, which means the value was lower than the limits to support larvae survival.
Conclusion
The application of probiotics containing B. subtilis in the mud crab, S. tranquebarica larvae rearing at the doses of 5 mg/L to 7.5 mg/L with the application started on day seventh of larval rearing with intervals every three days successfully to suppress Vibrio spp. development and improving water quality in larvae rearing, furthermore its impacts to the higher larvae survival rate and finally can improve S. tranquebarica crablet production significantly.







